Tutorial: Running the pipeline
Source:vignettes/vignette_run_pipeline.Rmd
vignette_run_pipeline.RmdIntroduction
staRgate is an automated gating pipeline to process and analyze flow cytometry data to characterize the lineage, differentiation and functional states of T-cells.
This pipeline is designed to mimic the manual gating strategy of
defining flow biomarker positive populations relative to a unimodal
background population to include cells with varying intensities of
marker expression. This is achieved via estimating the kernel density of
the intensity distribution and corresponding derivatives. This pipeline
integrates the density gating method in conjuction with some
pre-processing steps achieved via the R package openCyto
and an optional step with flowAI.
The flow data is stored within R as a GatingSet object,
which makes it easily transferable to other flow cytometry workflows
available on BioConductor.
This vignette will walk through how to run the {staRgate} pipeline starting from importing an flow cytometry standard (FCS) file into R to preprocessing and gating, as well as identifying T-cell subpopulations for downstream analysis. This pipeline returns results at the single-cell level as well as the summarized sample-level data (for the percentages of positively expressing cells when identifying the subpopulations).
For illustration purposes, the example FCS file used in this vignette and stored in the package data is a concatenated file limited to the first 30k events/cells acquired to reduce the run time and file size.
After running {staRgate} to gate flow cytometry data, it is
recommended to perform some quality checks (QC) on the gate placements
to ensure they are reasonable. We suggest to use ridgeplots in addition
to the ggcyto::autoplot to visualize the density
distributions per marker across samples. When examining a large batch of
samples, downsampling, such as to a random sample of 10k CD3+ cells,
will make the QC process more manageable. In addition, random spot
checks of a few samples would also be helpful QC to detect any edge
cases.
Currently in this tutorial, we do not extend to the QC steps and suggest to lean on other examples for how to put together a ridgeplot for example. In the near future, we hope to incorporate some examples for the additional QC steps as well, stay tuned!
Installation
The {staRgate} package relies on a few Biocondcutor R packages. Before installing {staRgate}, first setup Bioconductor and install all dependencies.
Follow instructions here for installing Bioconductor
The essential dependencies for running the {staRgate} package include: flowCore, and flowWorkspace.
The following packages are needed to fully run the pipeline as shown in this Tutorial, but not required for the {staRgate} code chunks to run: openCyto, ggplot2, ggcyto, gt
# Load libraries
library(staRgate)
library(openCyto)
library(flowWorkspace)
#> As part of improvements to flowWorkspace, some behavior of
#> GatingSet objects has changed. For details, please read the section
#> titled "The cytoframe and cytoset classes" in the package vignette:
#>
#> vignette("flowWorkspace-Introduction", "flowWorkspace")
library(flowCore)
# Just for plotting in the vignette
library(ggplot2)
library(ggcyto)
#> Loading required package: ncdfFlow
#> Loading required package: BH
# Set up dynamic variables
pt_samp_nm <- "flow_sample_1"
## File path to the FCS file
path_fcs <- system.file("extdata", "example_fcs.fcs", package = "staRgate", mustWork = TRUE)
## File path to the compensation matrix csv file
## Expect format to match flowJo exported version
path_comp_mat <- system.file("extdata", "comp_mat_example_fcs.csv", package = "staRgate", mustWork = TRUE)
## File path for outputs/saving
# Maybe not the best sol, but create a temp dir?
path_out <- tempdir()
# Print the path_out for user to see
path_out
#> [1] "/tmp/Rtmp3ZtP67"
## File path Gating template
gtFile <- system.file("extdata", "gating_template_x50_tcell.csv", package = "staRgate", mustWork = TRUE)
## File path to biexp parameters
## Expects 4 columns: full_name, ext_neg_dec, width_basis, positive_dec
## full name should contain the channel/dye name
# 3 remaining cols fill in with desired parameter values
path_biexp_params <- system.file("extdata", "biexp_transf_parameters_x50.csv", package = "staRgate", mustWork = TRUE)
## File path to positive peak thresholds
path_pos_peak_thresholds <- system.file("extdata", "pos_peak_thresholds.csv", package = "staRgate", mustWork = TRUE)Input files and function parameters
In order to run the pipeline, the user must have the data in flow
cytometry standard (FCS) format. This is usually the output from flowJo.
All of these input files except the bin size are expected
to be comma-separated values (csv) files. Please see the
inst folder of the package for examples of the formats.
- Compensation matrix from manual gating-
- Matrix where the column and row names correspond to the channel names, cell values correspond to the spillover correction to be applied.
- This can be exported as a csv in flowJo’s options
- Biexponential transformation parameters-
- This is a table specifying the parameters (negative decades, width basis and positive decades) to be applied to the listed channels
- Gating template
- A gating template is required to run the pre-gating via openCyto
- The package includes a gating template tailored for gating this panel of T-cell markers.
- For examples of how to modify the gating template, please refer to the openCyto documentation.
- Bin size
- From a systematic grid search, we have found bin sizes of 40 to 50 works well for the density gating
A few things to keep in mind when debugging/iterating through gating:
- If saving at the same path with same name (i.e., rerunning the same
code), the GatingSet folder from the
flowWorkspace::save_gs()command needs to be deleted for openCyto to save again, otherwise, will encounter an error related to an invalid path from theflowWorkspace::save_gs()function
Example
Below is an example of gating 1 FCS sample.
Import FCS
# Read in gating template
dtTemplate <- data.table::fread(gtFile)
# Load the FCS
gt_tcell <- openCyto::gatingTemplate(gtFile)
#> expanding pop: -/++/-
#> Adding population:fsc_ssc_qc
#> Adding population:nonDebris
#> Adding population:singlets
#> Adding population:cd14-cd19-
#> Adding population:live
#> Adding population:cd3
#> Adding population:cd4+
#> Adding population:cd8+
#> Adding population:cd4+cd8+
#> Adding population:cd4-cd8+
#> Adding population:cd4+cd8-
#> Adding population:cd4-cd8-
cs <- flowWorkspace::load_cytoset_from_fcs(path_fcs)
# Create a GatingSet of 1 sample
gs <- flowWorkspace::GatingSet(cs)
# Check- how many cells is in the FCS file?
n_root <- flowWorkspace::gh_pop_get_count(gs, "root")
# The example FCS has 30000 cells
n_root
#> [1] 30000Compensation
# Apply comp
gs <- getCompGS(gs, path_comp_mat = path_comp_mat)
# Can check that the comp was applied
chk_cm <- flowWorkspace::gh_get_compensations(gs)
# Not aware of an accessor that we can use for this
head(methods::slot(chk_cm, "spillover"), 2)
#> AF700-A APC-A APC-f750-A BB515-A BB660-A BB700-A
#> AF700-A 1.000000 0.0248301 0.2492840 0.009679220 0.00268438 0.04732480
#> APC-A 0.132334 1.0000000 0.0337338 -0.000160633 0.01598550 0.00443383
#> BB790-A BUV395-A BUV496-A BUV563-A BUV615-A BUV661-A
#> AF700-A 0.01730770 0.001813870 1.81722e-03 1.45041e-03 0.00249455 0.00220337
#> APC-A 0.00122597 0.000471851 -5.18352e-05 -3.25396e-05 0.00127167 0.11436300
#> BUV737-A BUV805-A BV421-A BV480-A BV510-A BV570-A
#> AF700-A 0.1387160 0.03310420 5.11994e-05 0.003390200 2.60901e-03 2.45899e-03
#> APC-A 0.0221613 0.00527436 -4.09868e-04 0.000113668 4.37466e-05 5.85607e-05
#> BV605-A BV650-A BV711-A BV750-A BV786-A PE-A
#> AF700-A 0.00358515 0.00445768 0.2127300 0.08483990 0.04394760 0.006840740
#> APC-A 0.00085079 0.13844100 0.0285134 0.00973321 0.00417643 0.000280757
#> PE-CF594-A PE-Cy5-A PE-Cy5.5-A PE-Cy7-A
#> AF700-A 0.02926360 0.0164829 0.3260510 0.1032330
#> APC-A 0.00541964 0.2677470 0.0810869 0.0215633Transformation
The transformation applied to all channels is the same: biexponential
with extra negative decades = 0.5,
positive decades = 4.5 and
width basis = -30
The structure of the table of parameters (as .csv format) should be first column for the flurochrome names corresponding to the panel, followed by the parameters.
tbl_biexp_params <-
utils::read.csv(path_biexp_params) |>
janitor::clean_names(case="all_caps")
head(tbl_biexp_params, 2)
#> FULL_NAME EXT_NEG_DEC WIDTH_BASIS POSITIVE_DEC
#> 1 BUV395-A 0.5 -30 4.5
#> 2 BUV496-A 0.5 -30 4.5Currently, the package only supports biexponetial transformation for
all channels with the getBiexpTransformGS() function.
However, the user may choose to create a transformation list explicitly
if other transformations (e.g., archsin) are desired.
Note that the flowWorkspace package also allows for an automated transformation calculation “guessing” appropriate parameters. We chose to explicitly specify the biexponential transformation with fixed parameters for all channels to match the manual gating strategy used in flowJo for a more direct comparison of {staRgate} when benchmarking against the manual gating results.
# Save the pre-transformed data to compare ranges
# And check that transformation was applied
dat_pre_transform <-
flowWorkspace::gh_pop_get_data(gs) |>
flowCore::exprs()
# Apply biexp trans
gs <- getBiexpTransformGS(gs, path_biexp_params = path_biexp_params)
## **Optional**-- to check what pre-transformed data against post
# save the post-transformed data
dat_post_transform <-
flowWorkspace::gh_pop_get_data(gs) |>
flowCore::exprs()
## **Optional**-- to check that the transformation worked on all provided channels!
## Commented out for ease of length
# summary(dat_pre_transform)
# summary(dat_post_transform)Pre-gating
In this context, pre-gating is defined as gating from the root population (all cells acquired) up to key parent populations: CD3+, or CD4+/CD8+ subsets. Then we will gate each marker indpendently.
The flowAI step serves as a quality control (QC) to match the first Time gate step that is typically done in manual gating. It is possible, however, that the user may choose to skip this step if flowAI excludes too many cells.
The first step of the gating template is a QC step that is especially important to include if the user chooses to exclude the flowAI step.
In this tutorial, we will skip the flowAI step to ease the length.
# Pre-gating up to CD4/8+ with `r BiocStyle::Biocpkg("openCyto")`
## Set seed using today's date
set.seed(glue::glue({
format(Sys.Date(), format = "%Y%m%d")
}))
openCyto::gt_gating(gt_tcell, gs)
#> Gating for 'fsc_ssc_qc'
#> done!
#> done.
#> Gating for 'nonDebris'
#> done!
#> done.
#> Gating for 'singlets'
#> done!
#> done.
#> Gating for 'cd14-cd19-'
#> done!
#> done.
#> Gating for 'live'
#> done!
#> done.
#> Gating for 'cd3'
#> done!
#> done.
#> Gating for 'cd8+'
#> done!
#> done.
#> Gating for 'cd4+'
#> done!
#> done.
#> Population 'cd4-cd8-'
#> done.
#> Population 'cd4+cd8-'
#> done.
#> Population 'cd4-cd8+'
#> done.
#> Population 'cd4+cd8+'
#> done.
#> finished.
## Check autoplot
ggcyto::autoplot(gs[[1]])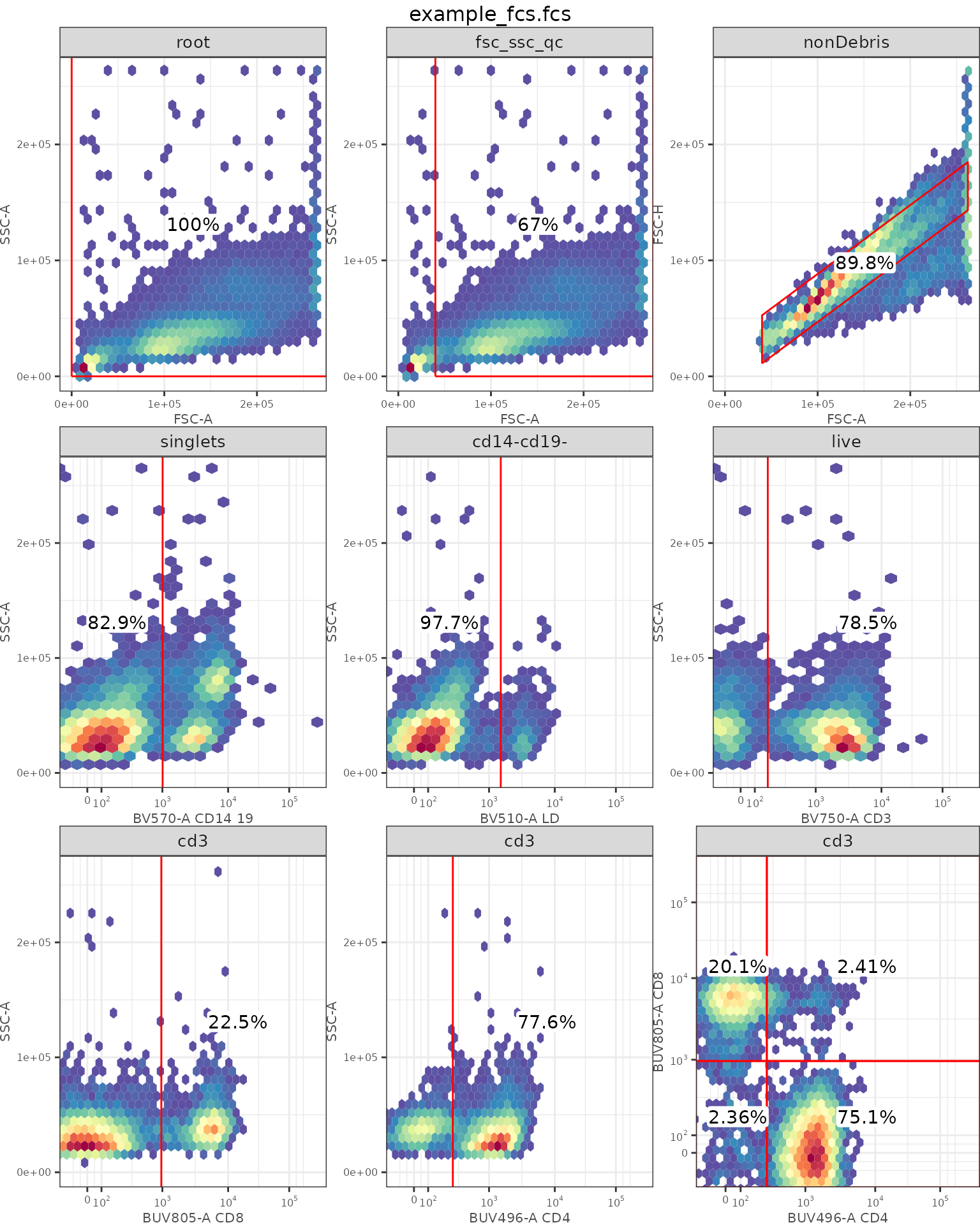
Extract intensity matrix
Grab the channel and marker names in the gs object
- When extracting
intensity_matrix, it is labeled with the channel names rather than the marker names. - The
marker_chnl_namesmapping created below will be used to rename the column names to the marker names, which will make calling the appropriate columns easier when analyzing the data
## Grab marker names from GatingSet for labeling col names in intensity matrix
## Can skip this step if you know the names of the channels that correspond to your marker names in your FCS files
# In that case, supply strings for `chnl` and `marker_full` is fine. Such as:
# chnl = c("BV750-A", "BUV496-A")
# marker_full = c("CD3", "CD4)
# This returns a named character vector
# With the channel names as names and marker names as the values
marker_chnl_names <- flowWorkspace::markernames(flowWorkspace::gh_pop_get_data(gs))
## Specify which markers to gate based on individual density distributions
# For our Tcell panel, we only want to apply the density gating on
# these 23 markers
markers_to_gate = c("CD45RA", "ICOS", "CD25", "TIM3",
"CD27", "CD57", "CXCR5", "CCR4",
"CCR7", "HLADR", "CD28", "PD1",
"LAG3", "CD127", "CD38", "TIGIT",
"EOMES", "CTLA4", "FOX_P3", "GITR",
"TBET", "KI67", "GZM_B")Next we grab the intensity values and indicators for the pre-gating steps.
# Extract intensity matrix from GatingSet object
## Grab the intensity matrix from GatingSet
intensity_dat <-
cbind(
# This grabs the intensity matrix with intensity values
flowWorkspace::gh_pop_get_data(gs) |>
flowCore::exprs(),
# the gh_pop_get_indices grabs the 0/1 for whether gated as pos/neg
# for each step specified
"fsc_ssc_qc" = flowWorkspace::gh_pop_get_indices(gs, y = "fsc_ssc_qc"),
"nonDebris" = flowWorkspace::gh_pop_get_indices(gs, y = "nonDebris"),
"singlets" = flowWorkspace::gh_pop_get_indices(gs, y = "singlets"),
"cd14_neg_19_neg" = flowWorkspace::gh_pop_get_indices(gs, y = "cd14-cd19-"),
"live" = flowWorkspace::gh_pop_get_indices(gs, y = "live"),
"cd3_pos" = flowWorkspace::gh_pop_get_indices(gs, y = "cd3"),
"cd4_pos" = flowWorkspace::gh_pop_get_indices(gs, y = "cd4+"),
"cd8_pos" = flowWorkspace::gh_pop_get_indices(gs, y = "cd8+")
) |>
# The intensity matrix is a matrix object. Convert to tibble.
tibble::as_tibble() |>
# Rename with colnames which are the channel names
# to marker names because it's easier to call columns by markers
# Rename using the marker_chnl_names we created above
# But we need it flipped when supplying to dplyr::rename()
# Where the names = value to rename to (marker names)
# The values of the vector = current names (Channel names)
dplyr::rename(
stats::setNames(names(marker_chnl_names),
# Clean up the marker names, make them all caps
janitor::make_clean_names(marker_chnl_names,
case = "all_caps",
replace = c("-" = "", "_" = "", " " = "")))
) |>
dplyr::mutate(
# Create a 4-level category for cd4, cd8 neg/pos
# in order to calculate the percentages individually within these parent populations
cd4_pos_cd8_pos = dplyr::case_when(
cd3_pos == 1 & cd4_pos == 1 & cd8_pos == 1 ~ "cd4_pos_cd8_pos",
cd3_pos == 1 & cd4_pos == 1 & cd8_pos == 0 ~ "cd4_pos_cd8_neg",
cd3_pos == 1 & cd4_pos == 0 & cd8_pos == 1 ~ "cd4_neg_cd8_pos",
cd3_pos == 1 & cd4_pos == 0 & cd8_pos == 0 ~ "cd4_neg_cd8_neg"
)
)
## Preview of intensity matrix
head(intensity_dat, 2)
#> # A tibble: 2 × 44
#> `FSC-A` `FSC-H` `FSC-W` `SSC-A` `SSC-H` `SSC-W` KI67 TBET GZM_B PD1 LAG3
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 14579. 12925. 91156. 6783. 6899. 59440. 898. 995. 949. 922. 1184.
#> 2 47535. 36802. 124831. 21010. 20507. 79210. 732. 1178. 775. 914. 1169.
#> # ℹ 33 more variables: CD127 <dbl>, CD38 <dbl>, CD45RA <dbl>, CD4 <dbl>,
#> # ICOS <dbl>, CD25 <dbl>, TIM3 <dbl>, CD27 <dbl>, CD8 <dbl>, CD57 <dbl>,
#> # CXCR5 <dbl>, LD <dbl>, CD1419 <dbl>, CCR4 <dbl>, CCR7 <dbl>, HLADR <dbl>,
#> # CD3 <dbl>, CD28 <dbl>, TIGIT <dbl>, EOMES <dbl>, CTLA4 <dbl>, FOX_P3 <dbl>,
#> # GITR <dbl>, Time <dbl>, fsc_ssc_qc <dbl>, nonDebris <dbl>, singlets <dbl>,
#> # cd14_neg_19_neg <dbl>, live <dbl>, cd3_pos <dbl>, cd4_pos <dbl>,
#> # cd8_pos <dbl>, cd4_pos_cd8_pos <chr>Gating on T-cell subsets
Pseudo-negative control
For this T-cell panel, we recommend using the CD3- cells as a pseudo-negative control when gating CD127 and CD28 as these two markers are expected to be predominantly negatively-expressing on CD3- cells. This mimics the isotype-based gating approach. Here we recommend using the 95th percentile of the CD3- distributions.
To ensure we only capture the CD3- cells, we first filter to the live
cells then to cd3_pos == 0
Empirical gating
For the other functional and differentiation markers where we cannot borrow CD3- as the pseudo-negative control, we gate empirically based on the CD3+ density distribution per marker.
- The suggested strategy is based on all CD3+, but this can be
customized based on the string corresponding to the column name supplied
to
subset_colin theget_density_gatesfunction - The suggested number of bins for density estimation is
40as some level of smoothing is required to reduce the noise and picking up false peaks. - When multiple samples from the same batch or experiment run (e.g., samples are processed on the same day), we recommend to borrow information from other samples by pooling all CD3+ across the same batch before applying the density gating.
- If interested in pooling across experiment runs, consider visualizing for any batch-level intensity shifts and variations before pooling samples together.
For illustration purposes, we will only apply density gating on a few markers.
# Density gating parameters
# peak detection ratio where any peak < 1/10 of the tallest peak will be
# considered as noise
peak_r <- 10
# smoothing to apply to the density estimation
# Using the default of 512 creates many little bumps/noise that are artifacts
# From a systematic grid search, we found the bin sizes of ~40-50 works best
bin_i <- 40
# Remove any very negative values that are artifacts from autogating
# -1000 on the biexp transformed scale corresponds to roughly -3300 on the original
# intensity scale so this is quite conservative.
neg_intensity_thres <- -1000
# select a few markers to gate
example_markers <- c("LAG3", "CCR7", "CD45RA")
# Read in positive peak thresholds
pos_thres <- utils::read.csv(path_pos_peak_thresholds) |>
janitor::clean_names(case = "all_caps")
# calculate the gates
dens_gates_pre <-
dplyr::filter(intensity_dat, cd3_pos == 1) |>
getDensityGates(
intens_dat = _,
marker = example_markers,
subset_col = "cd3_pos",
bin_n = bin_i,
peak_detect_ratio = peak_r,
pos_peak_threshold = pos_thres,
neg_intensity_threshold = neg_intensity_thres
)
# Since we apply density gating on CD3+ cells but
# Would like to calculate subpopulations with CD4+ and CD8+ as
# the starting parent population, we need to add corresponding rows
# to pass into getGatedDat()
dens_gates <-
# Stack the pseudo-neg gated markers and empirically gated markers
dplyr::bind_cols(dens_gates_pre, gates_pseudo_neg) |>
tibble::add_row() |>
tibble::add_row() |>
tibble::add_row() |>
dplyr::mutate(cd4_pos_cd8_pos = c("cd4_neg_cd8_neg", "cd4_pos_cd8_neg", "cd4_neg_cd8_pos", "cd4_pos_cd8_pos")) |>
tidyr::fill(-cd4_pos_cd8_pos, .direction = "down")
# View updated gates with the col for CD4/CD8
dens_gates
#> # A tibble: 4 × 7
#> cd3_pos LAG3 CCR7 CD45RA CD127 CD28 cd4_pos_cd8_pos
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 1 1634. 1768. 1486. 1327. 1251. cd4_neg_cd8_neg
#> 2 1 1634. 1768. 1486. 1327. 1251. cd4_pos_cd8_neg
#> 3 1 1634. 1768. 1486. 1327. 1251. cd4_neg_cd8_pos
#> 4 1 1634. 1768. 1486. 1327. 1251. cd4_pos_cd8_pos
# get indicator col
example_intensity_gated <-
getGatedDat(
# Only gate T-cells, which are CD3+
dplyr::filter(intensity_dat, cd3_pos == 1),
subset_col = "cd4_pos_cd8_pos",
cutoffs = dens_gates
)Visualizing the gates
LAG3 for all CD3+
# Plot the gate for visual
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(LAG3)) +
geom_vline(
data = dens_gates,
aes(xintercept = LAG3),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of LAG3 intensity on all CD3+. Gate identifed by {staRgate} in blue.")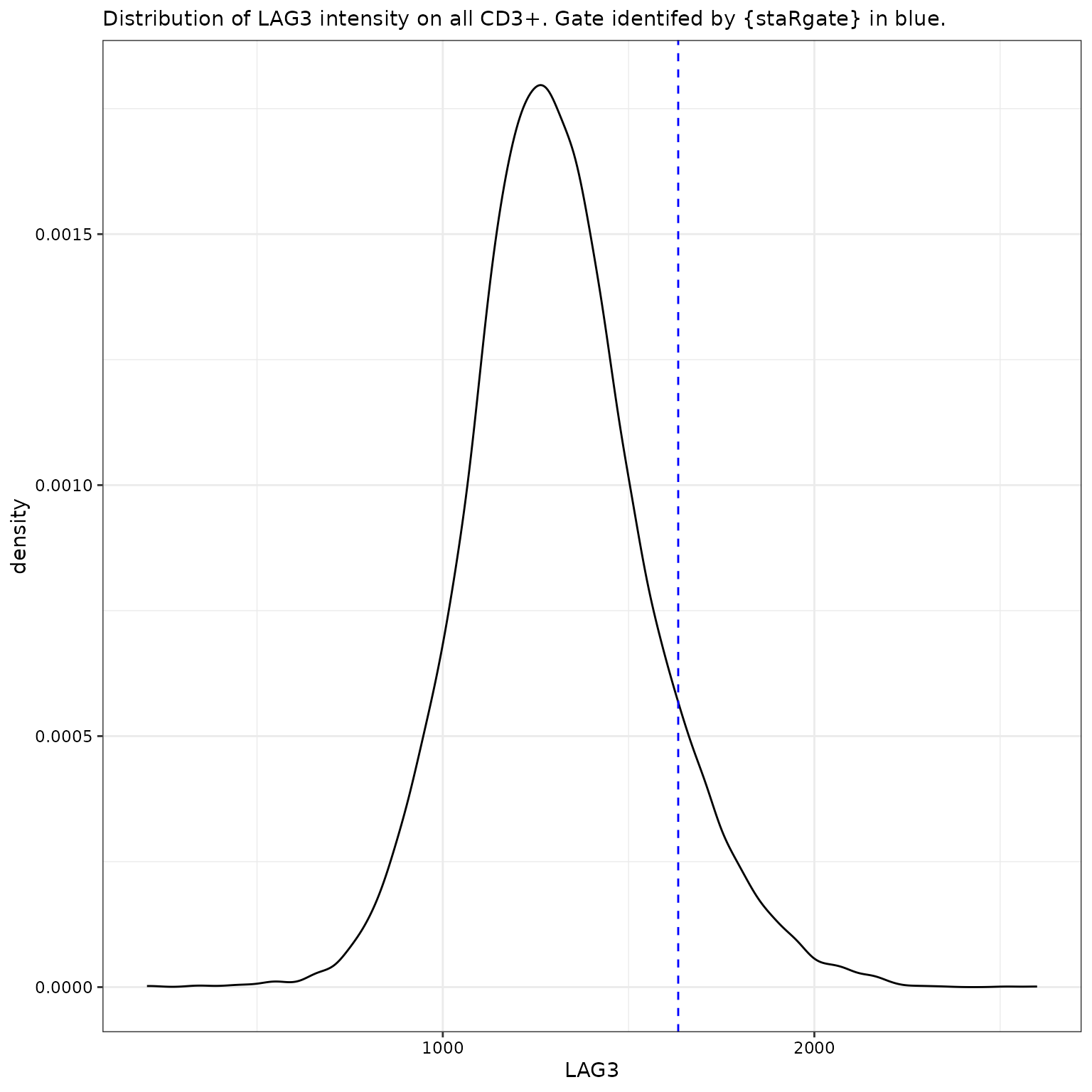
LAG3 by CD4 and CD8 subsets
# If by CD4/CD8,
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(LAG3)) +
geom_vline(
data = dens_gates,
aes(xintercept = LAG3),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of LAG3 intensity by CD4/CD8 subsets. Gate identifed by {staRgate} in blue.") +
facet_wrap(~cd4_pos_cd8_pos)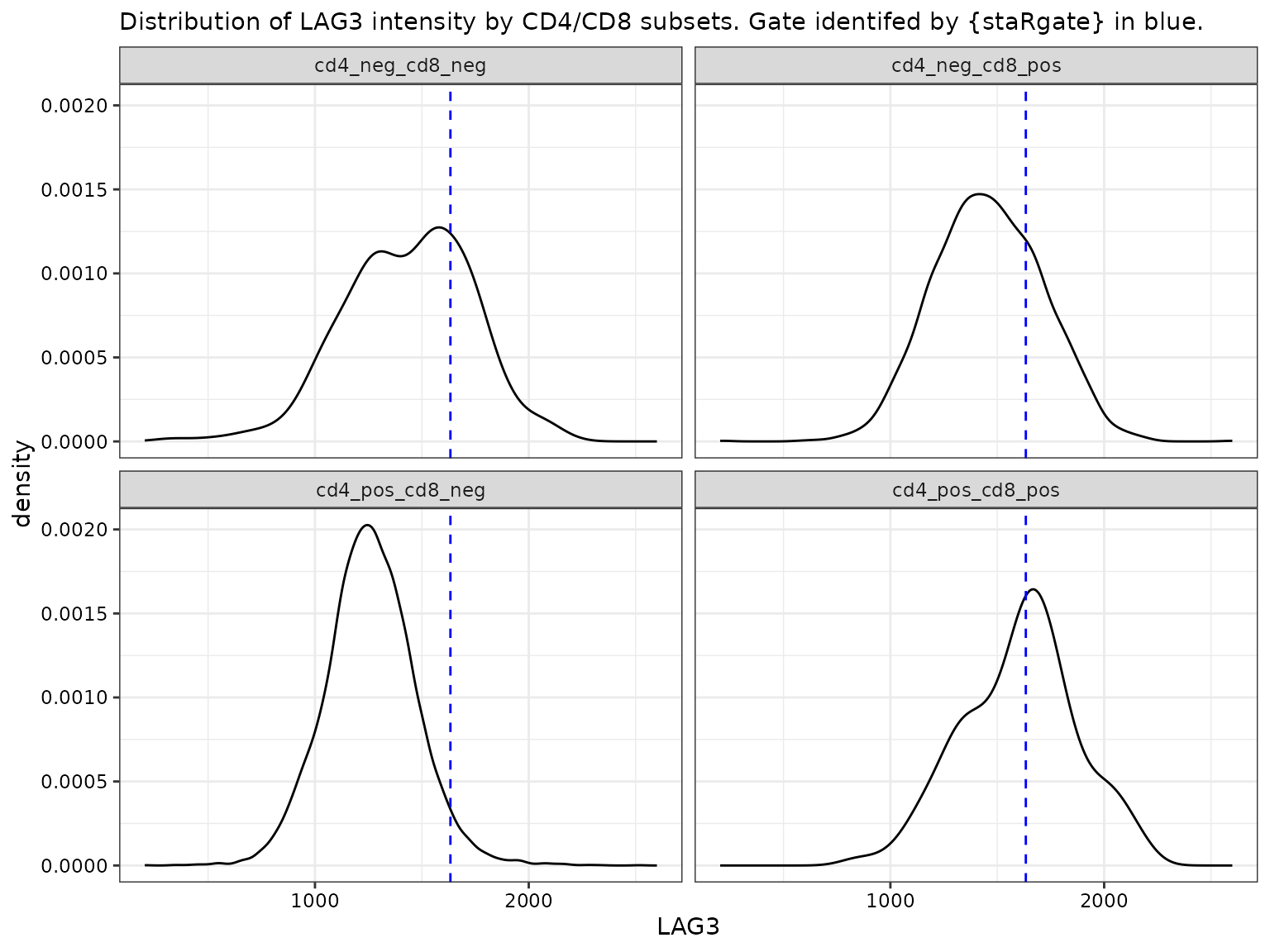
CCR7 for all CD3+
# For CCR7
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CCR7)) +
geom_vline(
data = dens_gates,
aes(xintercept = CCR7),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CCR7 intensity on all CD3+. Gate identifed by {staRgate} in blue.")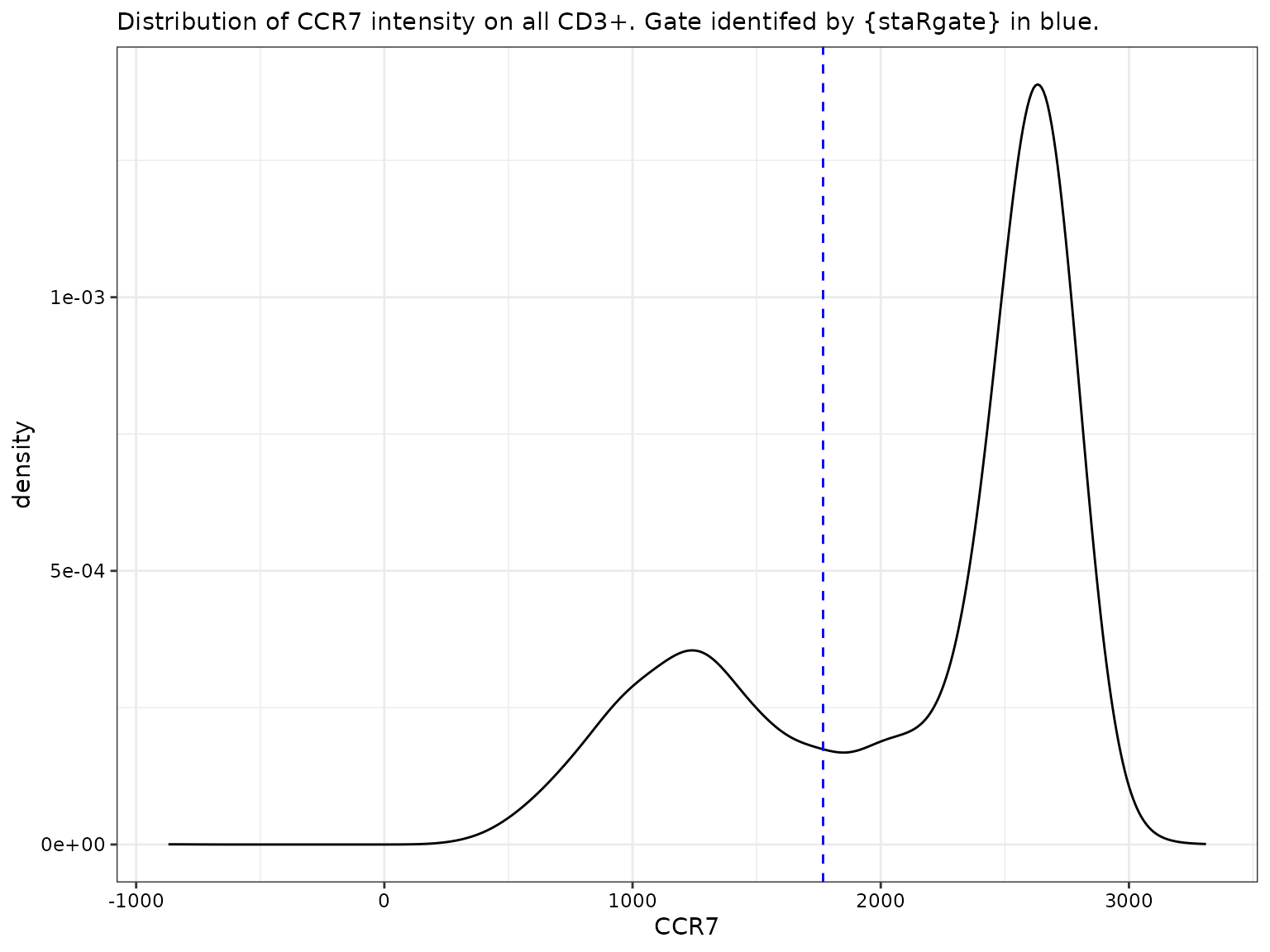
CCR7 by CD4 and CD8 subsets
# If by CD4/CD8,
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CCR7)) +
geom_vline(
data = dens_gates,
aes(xintercept = CCR7),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CCR7 intensity for CD4/CD8 subsets. Gate identifed by {staRgate} in blue.") +
facet_wrap(~cd4_pos_cd8_pos)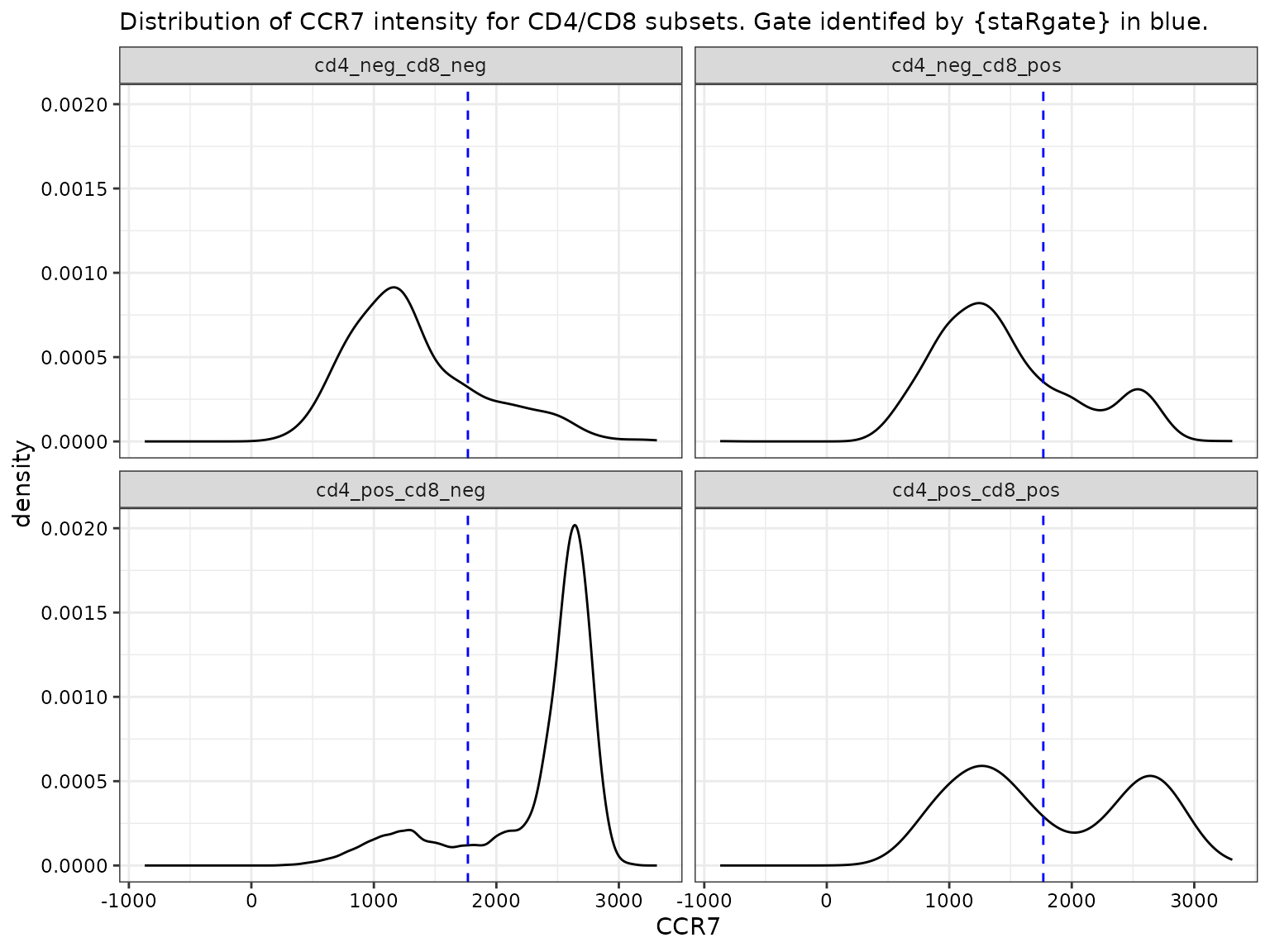
CD45RA for all CD3+
# For CD45RA
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD45RA)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD45RA),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD45RA intensity on all CD3+. Gate identifed by {staRgate} in blue.")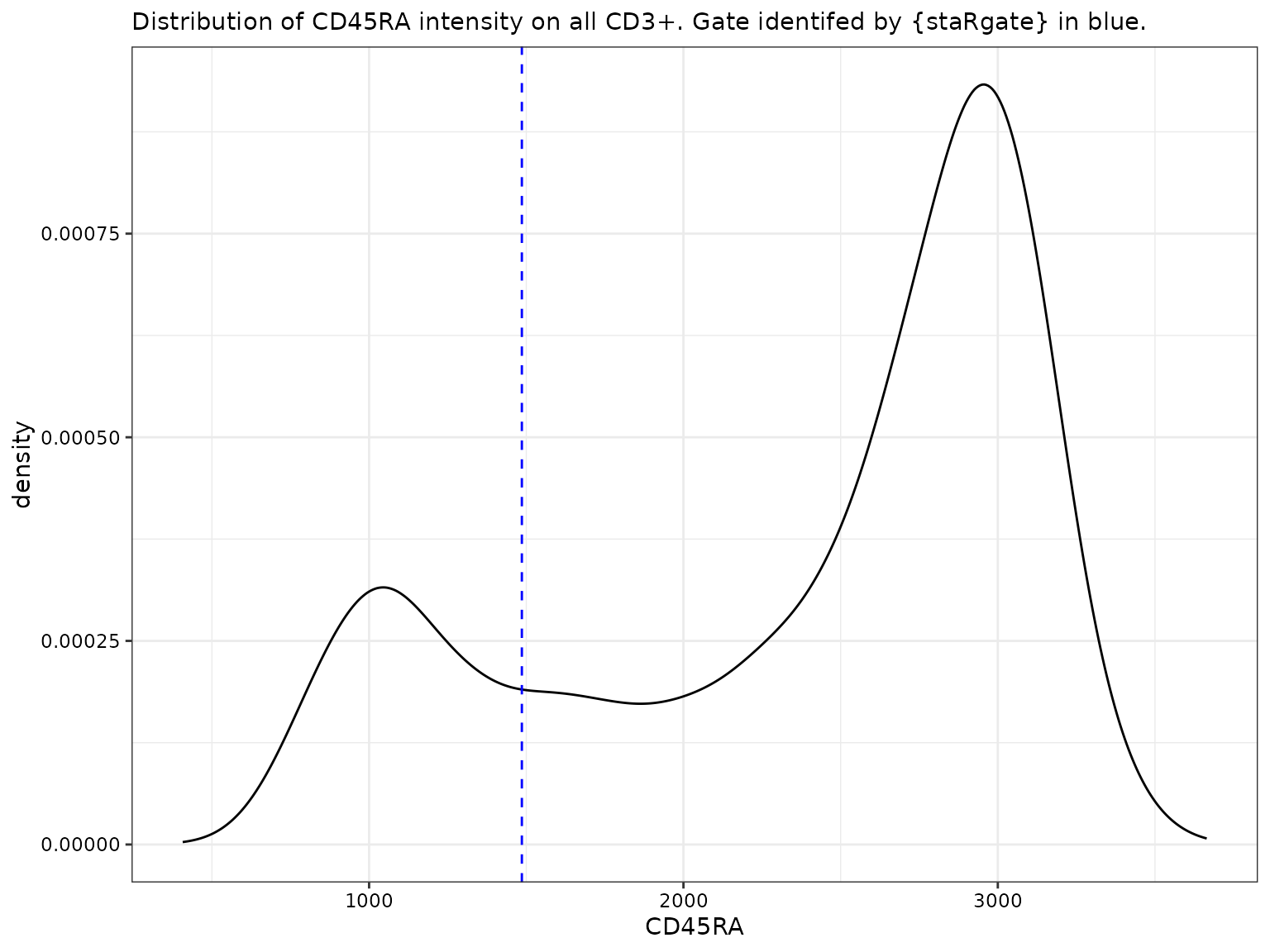
CD45RA by CD4 and CD8 subsets
# If by CD4/CD8,
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD45RA)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD45RA),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD45RA intensity for CD4/CD8 subsets. Gate identifed by {staRgate} in blue.") +
facet_wrap(~cd4_pos_cd8_pos)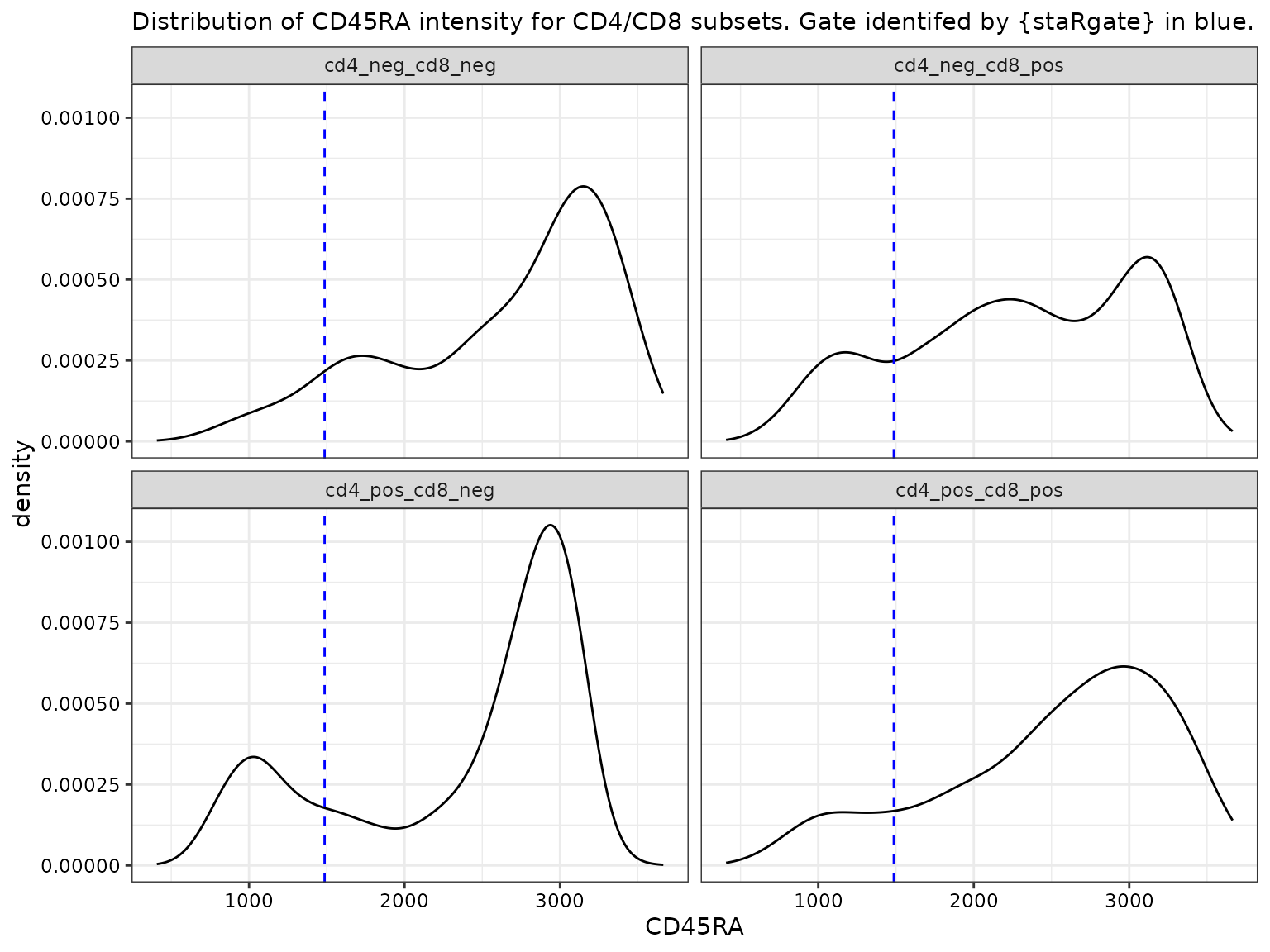
CD127 for all CD3+
# For CD127
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD127)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD127),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD127 intensity on all CD3+. Gate identifed by {staRgate} in blue.")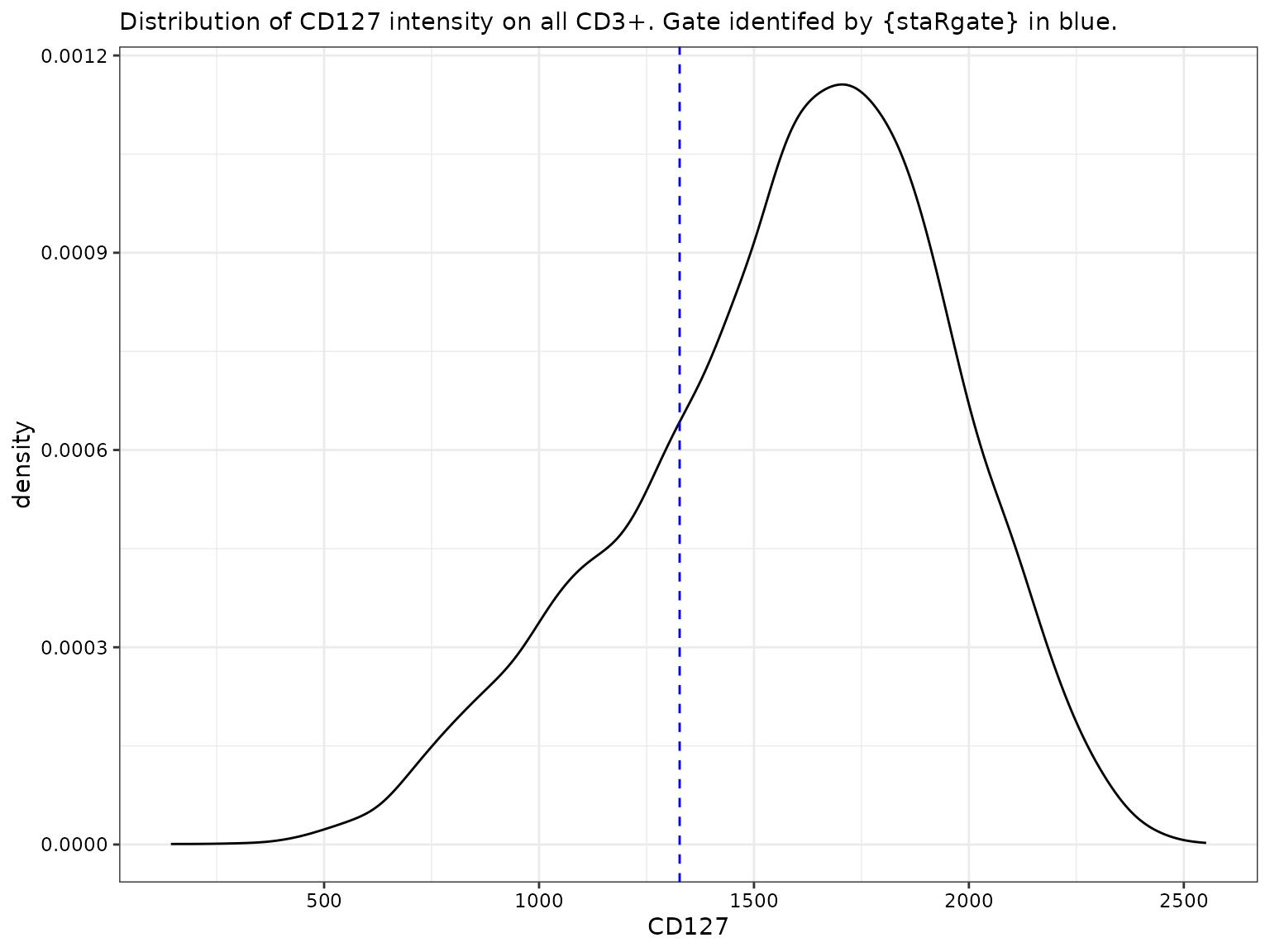
CD127 by CD4 and CD8 subsets
# If by CD4/CD8,
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD127)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD127),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD127 intensity for CD4/CD8 subsets. Gate identifed by {staRgate} in blue.") +
facet_wrap(~cd4_pos_cd8_pos)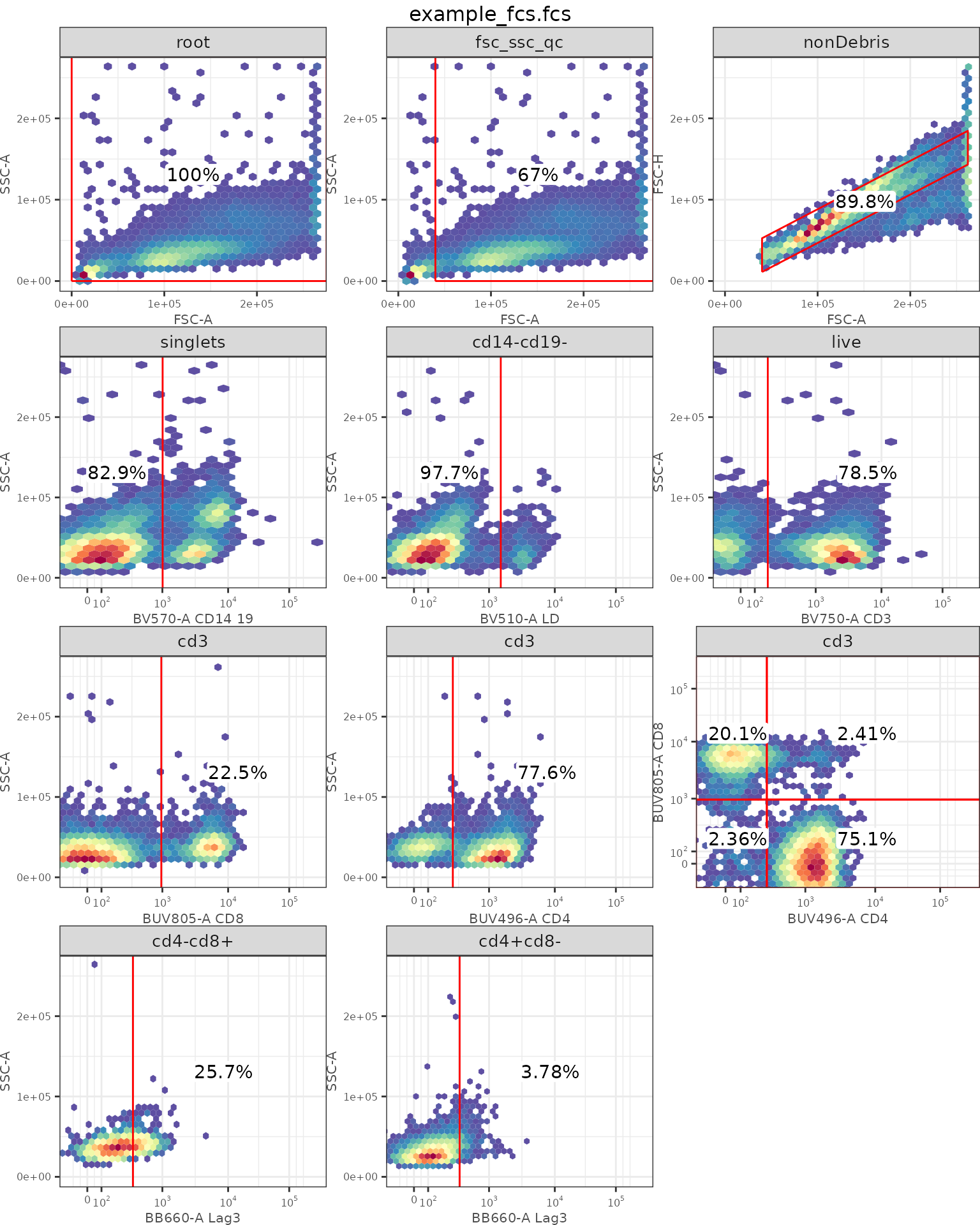
CD28 for all CD3+
# For CD28
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD28)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD28),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD28 intensity on all CD3+. Gate identifed by {staRgate} in blue.")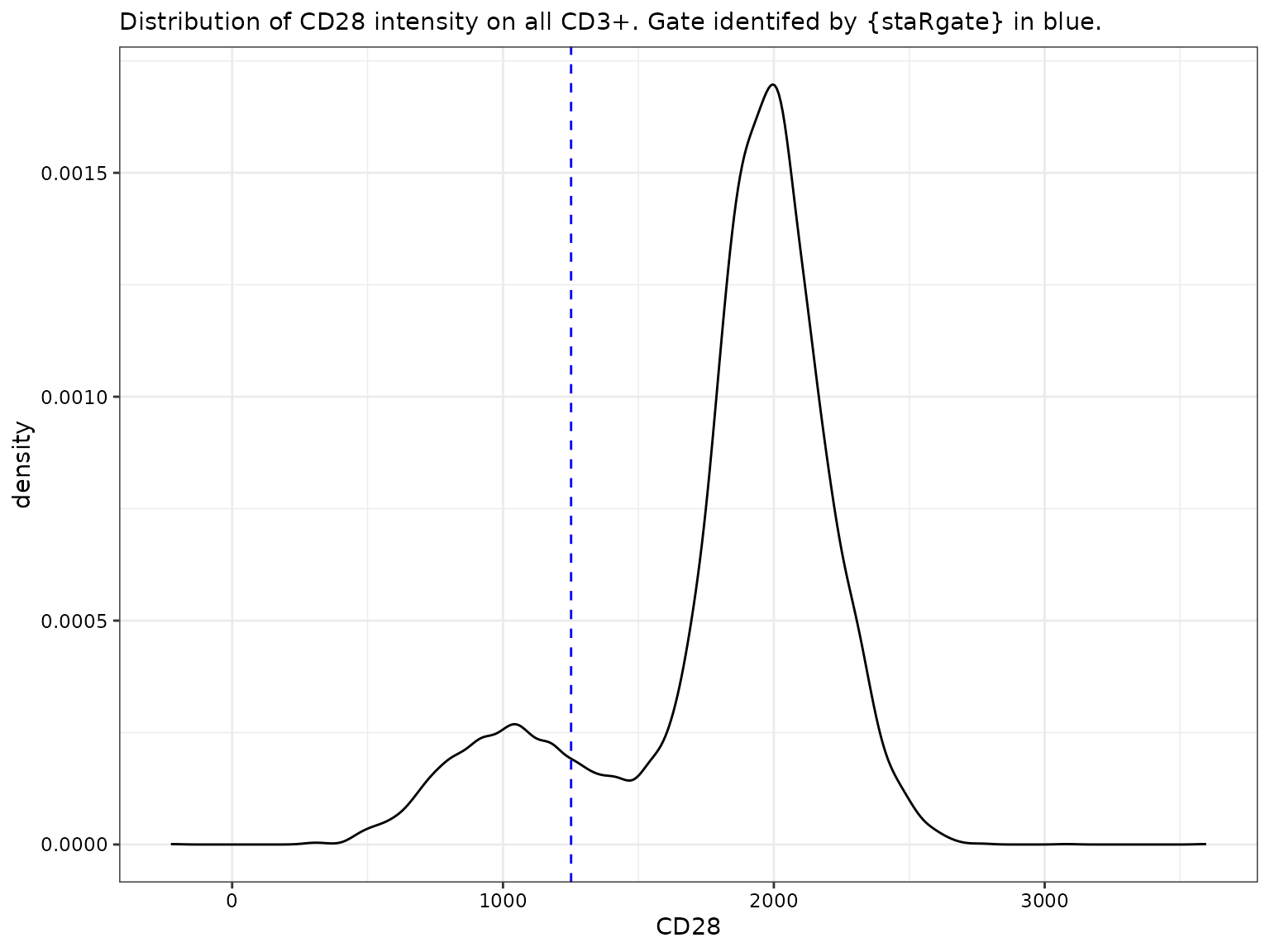
CD28 by CD4 and CD8 subsets
# If by CD4/CD8,
intensity_dat |>
dplyr::filter(cd3_pos == 1) |>
# additional step to remove large intensity values only when density gating.
# Still kept in the data
dplyr::filter(!(dplyr::if_any(dplyr::all_of(markers_to_gate), ~ .x < neg_intensity_thres))) |>
ggplot() +
geom_density(aes(CD28)) +
geom_vline(
data = dens_gates,
aes(xintercept = CD28),
color = "blue",
linetype = "dashed"
) +
labs(subtitle = "Distribution of CD28 intensity for CD4/CD8 subsets. Gate identifed by {staRgate} in blue.") +
facet_wrap(~cd4_pos_cd8_pos)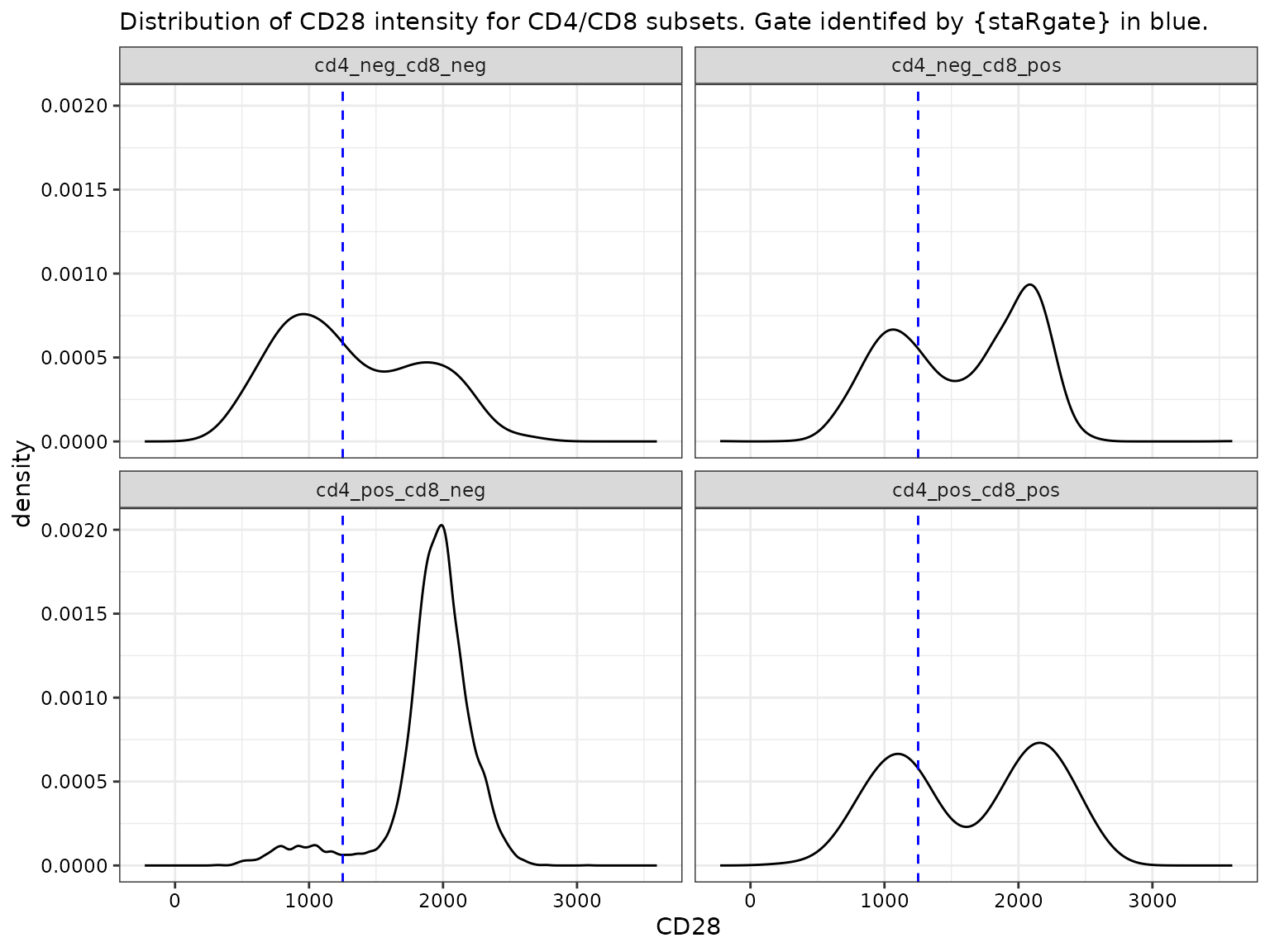
Getting percentage data
We can then summarize the single-cell level data to counts and percentages of cells for all combinations of markers.
For the subpopulations, the denominator is defined as
the parent population and numerator is the population of
interest out of the parent population. For example, the subpopulation
CD4+ of CD3+ cells correspond to the CD4+ as the numerator
and CD3+ as the denominator.
The refers to the number of markers considered for the denominator and for the number of markers considered for the numerator.
For the 29-marker panel, if the denominator is specified
as the CD4 and CD8 subsets, then
and
for the markers of interest.
The getPerc function allows user to list the markers of
interest for the numerator and denominator
In the example below, we will consider CD4 and CD8 subsets as the key
parent populations of interest (denominator) and the three
markers we gated on using c("LAG3", "CCR7", "CD45RA")
(numerator markers).
The additional arguments expand_num and
expand_denom generates different lists of subpopulations to
calculate counts/percentages for:
-
expand_num: should calculations consider up to pairs of numerator markers included?, -
expand_denom: should the calculations consider combinations of each numerator marker and parent populations specified in the denominator?
Currently, we support the four scenarios listed:
| expand_num | expand_denom | The subpopulations to generate | Examples | Expected number of subpopulations1 | Expected number of subpopulations for the 29-marker T-cell panel |
|---|---|---|---|---|---|
| FALSE | FALSE |
|
Numerator specified to (“LAG3”) and denominator specified as (“CD4”, “CD8”):
|
2n*(2nd) | 184 |
| TRUE | FALSE |
|
Numerator specified as (“LAG3”, “KI67”) and denominator as (“CD4”):
|
2n*(2nd) + 2nd*(n choose 2)*(22) |
4232 |
| FALSE | TRUE |
|
Numerator specified as (“LAG3”, “KI67”) and denominator as (“CD4”)
|
2n*(2nd) + 4*2nd*n*(n-1) |
8280 |
| TRUE | TRUE |
|
Numerator specified as (“LAG3”, “KI67”, “CTLA4”) and denominator as (“CD4”)
|
2n*(2nd) + 2nd*(n choose 2)*(22) + 4*2nd*n*(n-1) + 22*n*((n-1) choose 2)*(2nd+1) |
182344 |
| 1 n, number of markers specified in the numerator; nd, number of markers specified in the denominator | |||||
The keep_indicators argument provides the 0/1 for which
marker is considered in the numerator and denominator for each
subpopulation. This is especially useful when merging onto other data
that does not have the same naming conventions.
For example, when matching strings: “CD4+ & CD8- of CD3+” is different from “CD8- & CD4+ of CD3+” and “CD4+ and CD8- of CD3+”. But using indicator columns, we can match the subpopulations regardless of the subpopulation naming convention.
Below is we show examples of each of the four combinations for the
expand_num and expand_denom arguments.
For the example of when expand_num = FALSE and
expand_denom = FALSE, keep_indicators = TRUE
to illustrate the columns we get for the _POS and
_POS_D. Other examples use
keep_indicators = FALSE.
Expand example for expand_num = FALSE and
expand_denom = FALSE, and
keep_indicators = TRUE
example_perc1 <-
# Should only count the CD3+ cells
dplyr::filter(example_intensity_gated, cd3_pos == 1) |>
getPerc(
intens_dat = _,
num_marker = example_markers,
denom_marker = c("CD4", "CD8"),
expand_num = FALSE,
expand_denom = FALSE,
keep_indicators = TRUE
)
# For display only, group based on the denominators and
# simplify the names to be numerators
example_perc1 |>
tidyr::separate_wider_delim(subpopulation,
delim = "_OF_",
names = c("num", "denom"),
cols_remove = FALSE
) |>
dplyr::mutate(denom = paste("Denom = ", denom)) |>
dplyr::group_by(denom) |>
dplyr::select(-subpopulation) |>
gt::gt() |>
gt::fmt_number(
columns = "perc",
decimals = 1
)| num | n_num | n_denom | perc | LAG3_POS | CCR7_POS | CD45RA_POS | CD4_POS_D | CD8_POS_D |
|---|---|---|---|---|---|---|---|---|
| Denom = CD4_NEG_CD8_NEG | ||||||||
| LAG3_NEG | 196 | 271 | 72.3 | 0 | NA | NA | 0 | 0 |
| CCR7_NEG | 220 | 271 | 81.2 | NA | 0 | NA | 0 | 0 |
| CD45RA_NEG | 21 | 271 | 7.7 | NA | NA | 0 | 0 | 0 |
| LAG3_POS | 75 | 271 | 27.7 | 1 | NA | NA | 0 | 0 |
| CCR7_POS | 51 | 271 | 18.8 | NA | 1 | NA | 0 | 0 |
| CD45RA_POS | 250 | 271 | 92.3 | NA | NA | 1 | 0 | 0 |
| Denom = CD4_NEG_CD8_POS | ||||||||
| LAG3_NEG | 1711 | 2302 | 74.3 | 0 | NA | NA | 0 | 1 |
| CCR7_NEG | 1699 | 2302 | 73.8 | NA | 0 | NA | 0 | 1 |
| CD45RA_NEG | 413 | 2302 | 17.9 | NA | NA | 0 | 0 | 1 |
| LAG3_POS | 591 | 2302 | 25.7 | 1 | NA | NA | 0 | 1 |
| CCR7_POS | 603 | 2302 | 26.2 | NA | 1 | NA | 0 | 1 |
| CD45RA_POS | 1889 | 2302 | 82.1 | NA | NA | 1 | 0 | 1 |
| Denom = CD4_POS_CD8_NEG | ||||||||
| LAG3_NEG | 8289 | 8615 | 96.2 | 0 | NA | NA | 1 | 0 |
| CCR7_NEG | 1395 | 8615 | 16.2 | NA | 0 | NA | 1 | 0 |
| CD45RA_NEG | 1830 | 8615 | 21.2 | NA | NA | 0 | 1 | 0 |
| LAG3_POS | 326 | 8615 | 3.8 | 1 | NA | NA | 1 | 0 |
| CCR7_POS | 7220 | 8615 | 83.8 | NA | 1 | NA | 1 | 0 |
| CD45RA_POS | 6785 | 8615 | 78.8 | NA | NA | 1 | 1 | 0 |
| Denom = CD4_POS_CD8_POS | ||||||||
| LAG3_NEG | 139 | 276 | 50.4 | 0 | NA | NA | 1 | 1 |
| CCR7_NEG | 152 | 276 | 55.1 | NA | 0 | NA | 1 | 1 |
| CD45RA_NEG | 35 | 276 | 12.7 | NA | NA | 0 | 1 | 1 |
| LAG3_POS | 137 | 276 | 49.6 | 1 | NA | NA | 1 | 1 |
| CCR7_POS | 124 | 276 | 44.9 | NA | 1 | NA | 1 | 1 |
| CD45RA_POS | 241 | 276 | 87.3 | NA | NA | 1 | 1 | 1 |
Expand example for expand_num = TRUE and
expand_denom = FALSE, and
keep_indicators = FALSE
example_perc2 <-
# Should only count the CD3+ cells
dplyr::filter(example_intensity_gated, cd3_pos == 1) |>
getPerc(
intens_dat = _,
num_marker = example_markers,
denom_marker = c("CD4", "CD8"),
expand_num = TRUE,
expand_denom = FALSE,
keep_indicators = FALSE
)
# For display only, group based on the denominators and
# simplify the names to be numerators
example_perc2 |>
tidyr::separate_wider_delim(subpopulation,
delim = "_OF_",
names = c("num", "denom"),
cols_remove = FALSE
) |>
dplyr::mutate(denom = paste("Denom = ", denom)) |>
dplyr::group_by(denom) |>
dplyr::select(-subpopulation) |>
gt::gt() |>
gt::fmt_number(
columns = "perc",
decimals = 1
)| num | n_num | n_denom | perc |
|---|---|---|---|
| Denom = CD4_NEG_CD8_NEG | |||
| LAG3_NEG | 196 | 271 | 72.3 |
| CCR7_NEG | 220 | 271 | 81.2 |
| CD45RA_NEG | 21 | 271 | 7.7 |
| LAG3_POS | 75 | 271 | 27.7 |
| CCR7_POS | 51 | 271 | 18.8 |
| CD45RA_POS | 250 | 271 | 92.3 |
| LAG3_NEG_CCR7_NEG | 147 | 271 | 54.2 |
| LAG3_NEG_CD45RA_NEG | 17 | 271 | 6.3 |
| LAG3_NEG_CCR7_POS | 49 | 271 | 18.1 |
| LAG3_NEG_CD45RA_POS | 179 | 271 | 66.1 |
| CCR7_NEG_CD45RA_NEG | 17 | 271 | 6.3 |
| CCR7_NEG_LAG3_POS | 73 | 271 | 26.9 |
| CCR7_NEG_CD45RA_POS | 203 | 271 | 74.9 |
| CD45RA_NEG_LAG3_POS | 4 | 271 | 1.5 |
| CD45RA_NEG_CCR7_POS | 4 | 271 | 1.5 |
| LAG3_POS_CCR7_POS | 2 | 271 | 0.7 |
| LAG3_POS_CD45RA_POS | 71 | 271 | 26.2 |
| CCR7_POS_CD45RA_POS | 47 | 271 | 17.3 |
| Denom = CD4_NEG_CD8_POS | |||
| LAG3_NEG | 1711 | 2302 | 74.3 |
| CCR7_NEG | 1699 | 2302 | 73.8 |
| CD45RA_NEG | 413 | 2302 | 17.9 |
| LAG3_POS | 591 | 2302 | 25.7 |
| CCR7_POS | 603 | 2302 | 26.2 |
| CD45RA_POS | 1889 | 2302 | 82.1 |
| LAG3_NEG_CCR7_NEG | 1165 | 2302 | 50.6 |
| LAG3_NEG_CD45RA_NEG | 344 | 2302 | 14.9 |
| LAG3_NEG_CCR7_POS | 546 | 2302 | 23.7 |
| LAG3_NEG_CD45RA_POS | 1367 | 2302 | 59.4 |
| CCR7_NEG_CD45RA_NEG | 329 | 2302 | 14.3 |
| CCR7_NEG_LAG3_POS | 534 | 2302 | 23.2 |
| CCR7_NEG_CD45RA_POS | 1370 | 2302 | 59.5 |
| CD45RA_NEG_LAG3_POS | 69 | 2302 | 3.0 |
| CD45RA_NEG_CCR7_POS | 84 | 2302 | 3.6 |
| LAG3_POS_CCR7_POS | 57 | 2302 | 2.5 |
| LAG3_POS_CD45RA_POS | 522 | 2302 | 22.7 |
| CCR7_POS_CD45RA_POS | 519 | 2302 | 22.5 |
| Denom = CD4_POS_CD8_NEG | |||
| LAG3_NEG | 8289 | 8615 | 96.2 |
| CCR7_NEG | 1395 | 8615 | 16.2 |
| CD45RA_NEG | 1830 | 8615 | 21.2 |
| LAG3_POS | 326 | 8615 | 3.8 |
| CCR7_POS | 7220 | 8615 | 83.8 |
| CD45RA_POS | 6785 | 8615 | 78.8 |
| LAG3_NEG_CCR7_NEG | 1221 | 8615 | 14.2 |
| LAG3_NEG_CD45RA_NEG | 1700 | 8615 | 19.7 |
| LAG3_NEG_CCR7_POS | 7068 | 8615 | 82.0 |
| LAG3_NEG_CD45RA_POS | 6589 | 8615 | 76.5 |
| CCR7_NEG_CD45RA_NEG | 927 | 8615 | 10.8 |
| CCR7_NEG_LAG3_POS | 174 | 8615 | 2.0 |
| CCR7_NEG_CD45RA_POS | 468 | 8615 | 5.4 |
| CD45RA_NEG_LAG3_POS | 130 | 8615 | 1.5 |
| CD45RA_NEG_CCR7_POS | 903 | 8615 | 10.5 |
| LAG3_POS_CCR7_POS | 152 | 8615 | 1.8 |
| LAG3_POS_CD45RA_POS | 196 | 8615 | 2.3 |
| CCR7_POS_CD45RA_POS | 6317 | 8615 | 73.3 |
| Denom = CD4_POS_CD8_POS | |||
| LAG3_NEG | 139 | 276 | 50.4 |
| CCR7_NEG | 152 | 276 | 55.1 |
| CD45RA_NEG | 35 | 276 | 12.7 |
| LAG3_POS | 137 | 276 | 49.6 |
| CCR7_POS | 124 | 276 | 44.9 |
| CD45RA_POS | 241 | 276 | 87.3 |
| LAG3_NEG_CCR7_NEG | 79 | 276 | 28.6 |
| LAG3_NEG_CD45RA_NEG | 26 | 276 | 9.4 |
| LAG3_NEG_CCR7_POS | 60 | 276 | 21.7 |
| LAG3_NEG_CD45RA_POS | 113 | 276 | 40.9 |
| CCR7_NEG_CD45RA_NEG | 19 | 276 | 6.9 |
| CCR7_NEG_LAG3_POS | 73 | 276 | 26.4 |
| CCR7_NEG_CD45RA_POS | 133 | 276 | 48.2 |
| CD45RA_NEG_LAG3_POS | 9 | 276 | 3.3 |
| CD45RA_NEG_CCR7_POS | 16 | 276 | 5.8 |
| LAG3_POS_CCR7_POS | 64 | 276 | 23.2 |
| LAG3_POS_CD45RA_POS | 128 | 276 | 46.4 |
| CCR7_POS_CD45RA_POS | 108 | 276 | 39.1 |
Expand example for expand_num = FALSE and
expand_denom = TRUE, and
keep_indicators = FALSE
example_perc3 <-
# Should only count the CD3+ cells
dplyr::filter(example_intensity_gated, cd3_pos == 1) |>
getPerc(
intens_dat = _,
num_marker = example_markers,
denom_marker = c("CD4", "CD8"),
expand_num = FALSE,
expand_denom = TRUE,
keep_indicators = FALSE
)
# For display only, group based on the denominators and
# simplify the names to be numerators
example_perc3 |>
tidyr::separate_wider_delim(subpopulation,
delim = "_OF_",
names = c("num", "denom"),
cols_remove = FALSE
) |>
dplyr::mutate(denom = paste("Denom = ", denom)) |>
dplyr::group_by(denom) |>
dplyr::select(-subpopulation) |>
gt::gt() |>
gt::fmt_number(
columns = "perc",
decimals = 1
)| num | n_num | n_denom | perc |
|---|---|---|---|
| Denom = CD4_NEG_CD8_NEG | |||
| LAG3_NEG | 196 | 271 | 72.3 |
| CCR7_NEG | 220 | 271 | 81.2 |
| CD45RA_NEG | 21 | 271 | 7.7 |
| LAG3_POS | 75 | 271 | 27.7 |
| CCR7_POS | 51 | 271 | 18.8 |
| CD45RA_POS | 250 | 271 | 92.3 |
| Denom = CD4_NEG_CD8_POS | |||
| LAG3_NEG | 1711 | 2302 | 74.3 |
| CCR7_NEG | 1699 | 2302 | 73.8 |
| CD45RA_NEG | 413 | 2302 | 17.9 |
| LAG3_POS | 591 | 2302 | 25.7 |
| CCR7_POS | 603 | 2302 | 26.2 |
| CD45RA_POS | 1889 | 2302 | 82.1 |
| Denom = CD4_POS_CD8_NEG | |||
| LAG3_NEG | 8289 | 8615 | 96.2 |
| CCR7_NEG | 1395 | 8615 | 16.2 |
| CD45RA_NEG | 1830 | 8615 | 21.2 |
| LAG3_POS | 326 | 8615 | 3.8 |
| CCR7_POS | 7220 | 8615 | 83.8 |
| CD45RA_POS | 6785 | 8615 | 78.8 |
| Denom = CD4_POS_CD8_POS | |||
| LAG3_NEG | 139 | 276 | 50.4 |
| CCR7_NEG | 152 | 276 | 55.1 |
| CD45RA_NEG | 35 | 276 | 12.7 |
| LAG3_POS | 137 | 276 | 49.6 |
| CCR7_POS | 124 | 276 | 44.9 |
| CD45RA_POS | 241 | 276 | 87.3 |
| Denom = CD4_NEG_CD8_NEG_LAG3_NEG | |||
| CCR7_NEG | 147 | 196 | 75.0 |
| CD45RA_NEG | 17 | 196 | 8.7 |
| CCR7_POS | 49 | 196 | 25.0 |
| CD45RA_POS | 179 | 196 | 91.3 |
| Denom = CD4_NEG_CD8_POS_LAG3_NEG | |||
| CCR7_NEG | 1165 | 1711 | 68.1 |
| CD45RA_NEG | 344 | 1711 | 20.1 |
| CCR7_POS | 546 | 1711 | 31.9 |
| CD45RA_POS | 1367 | 1711 | 79.9 |
| Denom = CD4_POS_CD8_NEG_LAG3_NEG | |||
| CCR7_NEG | 1221 | 8289 | 14.7 |
| CD45RA_NEG | 1700 | 8289 | 20.5 |
| CCR7_POS | 7068 | 8289 | 85.3 |
| CD45RA_POS | 6589 | 8289 | 79.5 |
| Denom = CD4_POS_CD8_POS_LAG3_NEG | |||
| CCR7_NEG | 79 | 139 | 56.8 |
| CD45RA_NEG | 26 | 139 | 18.7 |
| CCR7_POS | 60 | 139 | 43.2 |
| CD45RA_POS | 113 | 139 | 81.3 |
| Denom = CD4_NEG_CD8_NEG_CCR7_NEG | |||
| LAG3_NEG | 147 | 220 | 66.8 |
| CD45RA_NEG | 17 | 220 | 7.7 |
| LAG3_POS | 73 | 220 | 33.2 |
| CD45RA_POS | 203 | 220 | 92.3 |
| Denom = CD4_NEG_CD8_POS_CCR7_NEG | |||
| LAG3_NEG | 1165 | 1699 | 68.6 |
| CD45RA_NEG | 329 | 1699 | 19.4 |
| LAG3_POS | 534 | 1699 | 31.4 |
| CD45RA_POS | 1370 | 1699 | 80.6 |
| Denom = CD4_POS_CD8_NEG_CCR7_NEG | |||
| LAG3_NEG | 1221 | 1395 | 87.5 |
| CD45RA_NEG | 927 | 1395 | 66.5 |
| LAG3_POS | 174 | 1395 | 12.5 |
| CD45RA_POS | 468 | 1395 | 33.5 |
| Denom = CD4_POS_CD8_POS_CCR7_NEG | |||
| LAG3_NEG | 79 | 152 | 52.0 |
| CD45RA_NEG | 19 | 152 | 12.5 |
| LAG3_POS | 73 | 152 | 48.0 |
| CD45RA_POS | 133 | 152 | 87.5 |
| Denom = CD4_NEG_CD8_NEG_CD45RA_NEG | |||
| LAG3_NEG | 17 | 21 | 81.0 |
| CCR7_NEG | 17 | 21 | 81.0 |
| LAG3_POS | 4 | 21 | 19.0 |
| CCR7_POS | 4 | 21 | 19.0 |
| Denom = CD4_NEG_CD8_POS_CD45RA_NEG | |||
| LAG3_NEG | 344 | 413 | 83.3 |
| CCR7_NEG | 329 | 413 | 79.7 |
| LAG3_POS | 69 | 413 | 16.7 |
| CCR7_POS | 84 | 413 | 20.3 |
| Denom = CD4_POS_CD8_NEG_CD45RA_NEG | |||
| LAG3_NEG | 1700 | 1830 | 92.9 |
| CCR7_NEG | 927 | 1830 | 50.7 |
| LAG3_POS | 130 | 1830 | 7.1 |
| CCR7_POS | 903 | 1830 | 49.3 |
| Denom = CD4_POS_CD8_POS_CD45RA_NEG | |||
| LAG3_NEG | 26 | 35 | 74.3 |
| CCR7_NEG | 19 | 35 | 54.3 |
| LAG3_POS | 9 | 35 | 25.7 |
| CCR7_POS | 16 | 35 | 45.7 |
| Denom = CD4_NEG_CD8_NEG_LAG3_POS | |||
| CCR7_NEG | 73 | 75 | 97.3 |
| CD45RA_NEG | 4 | 75 | 5.3 |
| CCR7_POS | 2 | 75 | 2.7 |
| CD45RA_POS | 71 | 75 | 94.7 |
| Denom = CD4_NEG_CD8_POS_LAG3_POS | |||
| CCR7_NEG | 534 | 591 | 90.4 |
| CD45RA_NEG | 69 | 591 | 11.7 |
| CCR7_POS | 57 | 591 | 9.6 |
| CD45RA_POS | 522 | 591 | 88.3 |
| Denom = CD4_POS_CD8_NEG_LAG3_POS | |||
| CCR7_NEG | 174 | 326 | 53.4 |
| CD45RA_NEG | 130 | 326 | 39.9 |
| CCR7_POS | 152 | 326 | 46.6 |
| CD45RA_POS | 196 | 326 | 60.1 |
| Denom = CD4_POS_CD8_POS_LAG3_POS | |||
| CCR7_NEG | 73 | 137 | 53.3 |
| CD45RA_NEG | 9 | 137 | 6.6 |
| CCR7_POS | 64 | 137 | 46.7 |
| CD45RA_POS | 128 | 137 | 93.4 |
| Denom = CD4_NEG_CD8_NEG_CCR7_POS | |||
| LAG3_NEG | 49 | 51 | 96.1 |
| CD45RA_NEG | 4 | 51 | 7.8 |
| LAG3_POS | 2 | 51 | 3.9 |
| CD45RA_POS | 47 | 51 | 92.2 |
| Denom = CD4_NEG_CD8_POS_CCR7_POS | |||
| LAG3_NEG | 546 | 603 | 90.5 |
| CD45RA_NEG | 84 | 603 | 13.9 |
| LAG3_POS | 57 | 603 | 9.5 |
| CD45RA_POS | 519 | 603 | 86.1 |
| Denom = CD4_POS_CD8_NEG_CCR7_POS | |||
| LAG3_NEG | 7068 | 7220 | 97.9 |
| CD45RA_NEG | 903 | 7220 | 12.5 |
| LAG3_POS | 152 | 7220 | 2.1 |
| CD45RA_POS | 6317 | 7220 | 87.5 |
| Denom = CD4_POS_CD8_POS_CCR7_POS | |||
| LAG3_NEG | 60 | 124 | 48.4 |
| CD45RA_NEG | 16 | 124 | 12.9 |
| LAG3_POS | 64 | 124 | 51.6 |
| CD45RA_POS | 108 | 124 | 87.1 |
| Denom = CD4_NEG_CD8_NEG_CD45RA_POS | |||
| LAG3_NEG | 179 | 250 | 71.6 |
| CCR7_NEG | 203 | 250 | 81.2 |
| LAG3_POS | 71 | 250 | 28.4 |
| CCR7_POS | 47 | 250 | 18.8 |
| Denom = CD4_NEG_CD8_POS_CD45RA_POS | |||
| LAG3_NEG | 1367 | 1889 | 72.4 |
| CCR7_NEG | 1370 | 1889 | 72.5 |
| LAG3_POS | 522 | 1889 | 27.6 |
| CCR7_POS | 519 | 1889 | 27.5 |
| Denom = CD4_POS_CD8_NEG_CD45RA_POS | |||
| LAG3_NEG | 6589 | 6785 | 97.1 |
| CCR7_NEG | 468 | 6785 | 6.9 |
| LAG3_POS | 196 | 6785 | 2.9 |
| CCR7_POS | 6317 | 6785 | 93.1 |
| Denom = CD4_POS_CD8_POS_CD45RA_POS | |||
| LAG3_NEG | 113 | 241 | 46.9 |
| CCR7_NEG | 133 | 241 | 55.2 |
| LAG3_POS | 128 | 241 | 53.1 |
| CCR7_POS | 108 | 241 | 44.8 |
Expand example for expand_num = TRUE and
expand_denom = TRUE , and
keep_indicators = FALSE
example_perc4 <-
# Should only count the CD3+ cells
dplyr::filter(example_intensity_gated, cd3_pos == 1) |>
getPerc(
intens_dat = _,
num_marker = example_markers,
denom_marker = c("CD4", "CD8"),
expand_num = TRUE,
expand_denom = TRUE,
keep_indicators = FALSE
)
# For display only, group based on the denominators and
# simplify the names to be numerators
example_perc4 |>
tidyr::separate_wider_delim(subpopulation,
delim = "_OF_",
names = c("num", "denom"),
cols_remove = FALSE
) |>
dplyr::mutate(denom = paste("Denom = ", denom)) |>
dplyr::group_by(denom) |>
dplyr::select(-subpopulation) |>
gt::gt() |>
gt::fmt_number(
columns = "perc",
decimals = 1
)| num | n_num | n_denom | perc |
|---|---|---|---|
| Denom = CD4_NEG_CD8_NEG | |||
| LAG3_NEG | 196 | 271 | 72.3 |
| CCR7_NEG | 220 | 271 | 81.2 |
| CD45RA_NEG | 21 | 271 | 7.7 |
| LAG3_POS | 75 | 271 | 27.7 |
| CCR7_POS | 51 | 271 | 18.8 |
| CD45RA_POS | 250 | 271 | 92.3 |
| LAG3_NEG_CCR7_NEG | 147 | 271 | 54.2 |
| LAG3_NEG_CD45RA_NEG | 17 | 271 | 6.3 |
| LAG3_NEG_CCR7_POS | 49 | 271 | 18.1 |
| LAG3_NEG_CD45RA_POS | 179 | 271 | 66.1 |
| CCR7_NEG_CD45RA_NEG | 17 | 271 | 6.3 |
| CCR7_NEG_LAG3_POS | 73 | 271 | 26.9 |
| CCR7_NEG_CD45RA_POS | 203 | 271 | 74.9 |
| CD45RA_NEG_LAG3_POS | 4 | 271 | 1.5 |
| CD45RA_NEG_CCR7_POS | 4 | 271 | 1.5 |
| LAG3_POS_CCR7_POS | 2 | 271 | 0.7 |
| LAG3_POS_CD45RA_POS | 71 | 271 | 26.2 |
| CCR7_POS_CD45RA_POS | 47 | 271 | 17.3 |
| Denom = CD4_NEG_CD8_POS | |||
| LAG3_NEG | 1711 | 2302 | 74.3 |
| CCR7_NEG | 1699 | 2302 | 73.8 |
| CD45RA_NEG | 413 | 2302 | 17.9 |
| LAG3_POS | 591 | 2302 | 25.7 |
| CCR7_POS | 603 | 2302 | 26.2 |
| CD45RA_POS | 1889 | 2302 | 82.1 |
| LAG3_NEG_CCR7_NEG | 1165 | 2302 | 50.6 |
| LAG3_NEG_CD45RA_NEG | 344 | 2302 | 14.9 |
| LAG3_NEG_CCR7_POS | 546 | 2302 | 23.7 |
| LAG3_NEG_CD45RA_POS | 1367 | 2302 | 59.4 |
| CCR7_NEG_CD45RA_NEG | 329 | 2302 | 14.3 |
| CCR7_NEG_LAG3_POS | 534 | 2302 | 23.2 |
| CCR7_NEG_CD45RA_POS | 1370 | 2302 | 59.5 |
| CD45RA_NEG_LAG3_POS | 69 | 2302 | 3.0 |
| CD45RA_NEG_CCR7_POS | 84 | 2302 | 3.6 |
| LAG3_POS_CCR7_POS | 57 | 2302 | 2.5 |
| LAG3_POS_CD45RA_POS | 522 | 2302 | 22.7 |
| CCR7_POS_CD45RA_POS | 519 | 2302 | 22.5 |
| Denom = CD4_POS_CD8_NEG | |||
| LAG3_NEG | 8289 | 8615 | 96.2 |
| CCR7_NEG | 1395 | 8615 | 16.2 |
| CD45RA_NEG | 1830 | 8615 | 21.2 |
| LAG3_POS | 326 | 8615 | 3.8 |
| CCR7_POS | 7220 | 8615 | 83.8 |
| CD45RA_POS | 6785 | 8615 | 78.8 |
| LAG3_NEG_CCR7_NEG | 1221 | 8615 | 14.2 |
| LAG3_NEG_CD45RA_NEG | 1700 | 8615 | 19.7 |
| LAG3_NEG_CCR7_POS | 7068 | 8615 | 82.0 |
| LAG3_NEG_CD45RA_POS | 6589 | 8615 | 76.5 |
| CCR7_NEG_CD45RA_NEG | 927 | 8615 | 10.8 |
| CCR7_NEG_LAG3_POS | 174 | 8615 | 2.0 |
| CCR7_NEG_CD45RA_POS | 468 | 8615 | 5.4 |
| CD45RA_NEG_LAG3_POS | 130 | 8615 | 1.5 |
| CD45RA_NEG_CCR7_POS | 903 | 8615 | 10.5 |
| LAG3_POS_CCR7_POS | 152 | 8615 | 1.8 |
| LAG3_POS_CD45RA_POS | 196 | 8615 | 2.3 |
| CCR7_POS_CD45RA_POS | 6317 | 8615 | 73.3 |
| Denom = CD4_POS_CD8_POS | |||
| LAG3_NEG | 139 | 276 | 50.4 |
| CCR7_NEG | 152 | 276 | 55.1 |
| CD45RA_NEG | 35 | 276 | 12.7 |
| LAG3_POS | 137 | 276 | 49.6 |
| CCR7_POS | 124 | 276 | 44.9 |
| CD45RA_POS | 241 | 276 | 87.3 |
| LAG3_NEG_CCR7_NEG | 79 | 276 | 28.6 |
| LAG3_NEG_CD45RA_NEG | 26 | 276 | 9.4 |
| LAG3_NEG_CCR7_POS | 60 | 276 | 21.7 |
| LAG3_NEG_CD45RA_POS | 113 | 276 | 40.9 |
| CCR7_NEG_CD45RA_NEG | 19 | 276 | 6.9 |
| CCR7_NEG_LAG3_POS | 73 | 276 | 26.4 |
| CCR7_NEG_CD45RA_POS | 133 | 276 | 48.2 |
| CD45RA_NEG_LAG3_POS | 9 | 276 | 3.3 |
| CD45RA_NEG_CCR7_POS | 16 | 276 | 5.8 |
| LAG3_POS_CCR7_POS | 64 | 276 | 23.2 |
| LAG3_POS_CD45RA_POS | 128 | 276 | 46.4 |
| CCR7_POS_CD45RA_POS | 108 | 276 | 39.1 |
| Denom = CD4_NEG_CD8_NEG_LAG3_NEG | |||
| CCR7_NEG | 147 | 196 | 75.0 |
| CD45RA_NEG | 17 | 196 | 8.7 |
| CCR7_POS | 49 | 196 | 25.0 |
| CD45RA_POS | 179 | 196 | 91.3 |
| CCR7_NEG_CD45RA_NEG | 15 | 196 | 7.7 |
| CCR7_NEG_CD45RA_POS | 132 | 196 | 67.3 |
| CD45RA_NEG_CCR7_POS | 2 | 196 | 1.0 |
| CCR7_POS_CD45RA_POS | 47 | 196 | 24.0 |
| Denom = CD4_NEG_CD8_POS_LAG3_NEG | |||
| CCR7_NEG | 1165 | 1711 | 68.1 |
| CD45RA_NEG | 344 | 1711 | 20.1 |
| CCR7_POS | 546 | 1711 | 31.9 |
| CD45RA_POS | 1367 | 1711 | 79.9 |
| CCR7_NEG_CD45RA_NEG | 269 | 1711 | 15.7 |
| CCR7_NEG_CD45RA_POS | 896 | 1711 | 52.4 |
| CD45RA_NEG_CCR7_POS | 75 | 1711 | 4.4 |
| CCR7_POS_CD45RA_POS | 471 | 1711 | 27.5 |
| Denom = CD4_POS_CD8_NEG_LAG3_NEG | |||
| CCR7_NEG | 1221 | 8289 | 14.7 |
| CD45RA_NEG | 1700 | 8289 | 20.5 |
| CCR7_POS | 7068 | 8289 | 85.3 |
| CD45RA_POS | 6589 | 8289 | 79.5 |
| CCR7_NEG_CD45RA_NEG | 837 | 8289 | 10.1 |
| CCR7_NEG_CD45RA_POS | 384 | 8289 | 4.6 |
| CD45RA_NEG_CCR7_POS | 863 | 8289 | 10.4 |
| CCR7_POS_CD45RA_POS | 6205 | 8289 | 74.9 |
| Denom = CD4_POS_CD8_POS_LAG3_NEG | |||
| CCR7_NEG | 79 | 139 | 56.8 |
| CD45RA_NEG | 26 | 139 | 18.7 |
| CCR7_POS | 60 | 139 | 43.2 |
| CD45RA_POS | 113 | 139 | 81.3 |
| CCR7_NEG_CD45RA_NEG | 11 | 139 | 7.9 |
| CCR7_NEG_CD45RA_POS | 68 | 139 | 48.9 |
| CD45RA_NEG_CCR7_POS | 15 | 139 | 10.8 |
| CCR7_POS_CD45RA_POS | 45 | 139 | 32.4 |
| Denom = CD4_NEG_CD8_NEG_CCR7_NEG | |||
| LAG3_NEG | 147 | 220 | 66.8 |
| CD45RA_NEG | 17 | 220 | 7.7 |
| LAG3_POS | 73 | 220 | 33.2 |
| CD45RA_POS | 203 | 220 | 92.3 |
| LAG3_NEG_CD45RA_NEG | 15 | 220 | 6.8 |
| LAG3_NEG_CD45RA_POS | 132 | 220 | 60.0 |
| CD45RA_NEG_LAG3_POS | 2 | 220 | 0.9 |
| LAG3_POS_CD45RA_POS | 71 | 220 | 32.3 |
| Denom = CD4_NEG_CD8_POS_CCR7_NEG | |||
| LAG3_NEG | 1165 | 1699 | 68.6 |
| CD45RA_NEG | 329 | 1699 | 19.4 |
| LAG3_POS | 534 | 1699 | 31.4 |
| CD45RA_POS | 1370 | 1699 | 80.6 |
| LAG3_NEG_CD45RA_NEG | 269 | 1699 | 15.8 |
| LAG3_NEG_CD45RA_POS | 896 | 1699 | 52.7 |
| CD45RA_NEG_LAG3_POS | 60 | 1699 | 3.5 |
| LAG3_POS_CD45RA_POS | 474 | 1699 | 27.9 |
| Denom = CD4_POS_CD8_NEG_CCR7_NEG | |||
| LAG3_NEG | 1221 | 1395 | 87.5 |
| CD45RA_NEG | 927 | 1395 | 66.5 |
| LAG3_POS | 174 | 1395 | 12.5 |
| CD45RA_POS | 468 | 1395 | 33.5 |
| LAG3_NEG_CD45RA_NEG | 837 | 1395 | 60.0 |
| LAG3_NEG_CD45RA_POS | 384 | 1395 | 27.5 |
| CD45RA_NEG_LAG3_POS | 90 | 1395 | 6.5 |
| LAG3_POS_CD45RA_POS | 84 | 1395 | 6.0 |
| Denom = CD4_POS_CD8_POS_CCR7_NEG | |||
| LAG3_NEG | 79 | 152 | 52.0 |
| CD45RA_NEG | 19 | 152 | 12.5 |
| LAG3_POS | 73 | 152 | 48.0 |
| CD45RA_POS | 133 | 152 | 87.5 |
| LAG3_NEG_CD45RA_NEG | 11 | 152 | 7.2 |
| LAG3_NEG_CD45RA_POS | 68 | 152 | 44.7 |
| CD45RA_NEG_LAG3_POS | 8 | 152 | 5.3 |
| LAG3_POS_CD45RA_POS | 65 | 152 | 42.8 |
| Denom = CD4_NEG_CD8_NEG_CD45RA_NEG | |||
| LAG3_NEG | 17 | 21 | 81.0 |
| CCR7_NEG | 17 | 21 | 81.0 |
| LAG3_POS | 4 | 21 | 19.0 |
| CCR7_POS | 4 | 21 | 19.0 |
| LAG3_NEG_CCR7_NEG | 15 | 21 | 71.4 |
| LAG3_NEG_CCR7_POS | 2 | 21 | 9.5 |
| CCR7_NEG_LAG3_POS | 2 | 21 | 9.5 |
| LAG3_POS_CCR7_POS | 2 | 21 | 9.5 |
| Denom = CD4_NEG_CD8_POS_CD45RA_NEG | |||
| LAG3_NEG | 344 | 413 | 83.3 |
| CCR7_NEG | 329 | 413 | 79.7 |
| LAG3_POS | 69 | 413 | 16.7 |
| CCR7_POS | 84 | 413 | 20.3 |
| LAG3_NEG_CCR7_NEG | 269 | 413 | 65.1 |
| LAG3_NEG_CCR7_POS | 75 | 413 | 18.2 |
| CCR7_NEG_LAG3_POS | 60 | 413 | 14.5 |
| LAG3_POS_CCR7_POS | 9 | 413 | 2.2 |
| Denom = CD4_POS_CD8_NEG_CD45RA_NEG | |||
| LAG3_NEG | 1700 | 1830 | 92.9 |
| CCR7_NEG | 927 | 1830 | 50.7 |
| LAG3_POS | 130 | 1830 | 7.1 |
| CCR7_POS | 903 | 1830 | 49.3 |
| LAG3_NEG_CCR7_NEG | 837 | 1830 | 45.7 |
| LAG3_NEG_CCR7_POS | 863 | 1830 | 47.2 |
| CCR7_NEG_LAG3_POS | 90 | 1830 | 4.9 |
| LAG3_POS_CCR7_POS | 40 | 1830 | 2.2 |
| Denom = CD4_POS_CD8_POS_CD45RA_NEG | |||
| LAG3_NEG | 26 | 35 | 74.3 |
| CCR7_NEG | 19 | 35 | 54.3 |
| LAG3_POS | 9 | 35 | 25.7 |
| CCR7_POS | 16 | 35 | 45.7 |
| LAG3_NEG_CCR7_NEG | 11 | 35 | 31.4 |
| LAG3_NEG_CCR7_POS | 15 | 35 | 42.9 |
| CCR7_NEG_LAG3_POS | 8 | 35 | 22.9 |
| LAG3_POS_CCR7_POS | 1 | 35 | 2.9 |
| Denom = CD4_NEG_CD8_NEG_LAG3_POS | |||
| CCR7_NEG | 73 | 75 | 97.3 |
| CD45RA_NEG | 4 | 75 | 5.3 |
| CCR7_POS | 2 | 75 | 2.7 |
| CD45RA_POS | 71 | 75 | 94.7 |
| CCR7_NEG_CD45RA_NEG | 2 | 75 | 2.7 |
| CCR7_NEG_CD45RA_POS | 71 | 75 | 94.7 |
| CD45RA_NEG_CCR7_POS | 2 | 75 | 2.7 |
| CCR7_POS_CD45RA_POS | 0 | 75 | 0.0 |
| Denom = CD4_NEG_CD8_POS_LAG3_POS | |||
| CCR7_NEG | 534 | 591 | 90.4 |
| CD45RA_NEG | 69 | 591 | 11.7 |
| CCR7_POS | 57 | 591 | 9.6 |
| CD45RA_POS | 522 | 591 | 88.3 |
| CCR7_NEG_CD45RA_NEG | 60 | 591 | 10.2 |
| CCR7_NEG_CD45RA_POS | 474 | 591 | 80.2 |
| CD45RA_NEG_CCR7_POS | 9 | 591 | 1.5 |
| CCR7_POS_CD45RA_POS | 48 | 591 | 8.1 |
| Denom = CD4_POS_CD8_NEG_LAG3_POS | |||
| CCR7_NEG | 174 | 326 | 53.4 |
| CD45RA_NEG | 130 | 326 | 39.9 |
| CCR7_POS | 152 | 326 | 46.6 |
| CD45RA_POS | 196 | 326 | 60.1 |
| CCR7_NEG_CD45RA_NEG | 90 | 326 | 27.6 |
| CCR7_NEG_CD45RA_POS | 84 | 326 | 25.8 |
| CD45RA_NEG_CCR7_POS | 40 | 326 | 12.3 |
| CCR7_POS_CD45RA_POS | 112 | 326 | 34.4 |
| Denom = CD4_POS_CD8_POS_LAG3_POS | |||
| CCR7_NEG | 73 | 137 | 53.3 |
| CD45RA_NEG | 9 | 137 | 6.6 |
| CCR7_POS | 64 | 137 | 46.7 |
| CD45RA_POS | 128 | 137 | 93.4 |
| CCR7_NEG_CD45RA_NEG | 8 | 137 | 5.8 |
| CCR7_NEG_CD45RA_POS | 65 | 137 | 47.4 |
| CD45RA_NEG_CCR7_POS | 1 | 137 | 0.7 |
| CCR7_POS_CD45RA_POS | 63 | 137 | 46.0 |
| Denom = CD4_NEG_CD8_NEG_CCR7_POS | |||
| LAG3_NEG | 49 | 51 | 96.1 |
| CD45RA_NEG | 4 | 51 | 7.8 |
| LAG3_POS | 2 | 51 | 3.9 |
| CD45RA_POS | 47 | 51 | 92.2 |
| LAG3_NEG_CD45RA_NEG | 2 | 51 | 3.9 |
| LAG3_NEG_CD45RA_POS | 47 | 51 | 92.2 |
| CD45RA_NEG_LAG3_POS | 2 | 51 | 3.9 |
| LAG3_POS_CD45RA_POS | 0 | 51 | 0.0 |
| Denom = CD4_NEG_CD8_POS_CCR7_POS | |||
| LAG3_NEG | 546 | 603 | 90.5 |
| CD45RA_NEG | 84 | 603 | 13.9 |
| LAG3_POS | 57 | 603 | 9.5 |
| CD45RA_POS | 519 | 603 | 86.1 |
| LAG3_NEG_CD45RA_NEG | 75 | 603 | 12.4 |
| LAG3_NEG_CD45RA_POS | 471 | 603 | 78.1 |
| CD45RA_NEG_LAG3_POS | 9 | 603 | 1.5 |
| LAG3_POS_CD45RA_POS | 48 | 603 | 8.0 |
| Denom = CD4_POS_CD8_NEG_CCR7_POS | |||
| LAG3_NEG | 7068 | 7220 | 97.9 |
| CD45RA_NEG | 903 | 7220 | 12.5 |
| LAG3_POS | 152 | 7220 | 2.1 |
| CD45RA_POS | 6317 | 7220 | 87.5 |
| LAG3_NEG_CD45RA_NEG | 863 | 7220 | 12.0 |
| LAG3_NEG_CD45RA_POS | 6205 | 7220 | 85.9 |
| CD45RA_NEG_LAG3_POS | 40 | 7220 | 0.6 |
| LAG3_POS_CD45RA_POS | 112 | 7220 | 1.6 |
| Denom = CD4_POS_CD8_POS_CCR7_POS | |||
| LAG3_NEG | 60 | 124 | 48.4 |
| CD45RA_NEG | 16 | 124 | 12.9 |
| LAG3_POS | 64 | 124 | 51.6 |
| CD45RA_POS | 108 | 124 | 87.1 |
| LAG3_NEG_CD45RA_NEG | 15 | 124 | 12.1 |
| LAG3_NEG_CD45RA_POS | 45 | 124 | 36.3 |
| CD45RA_NEG_LAG3_POS | 1 | 124 | 0.8 |
| LAG3_POS_CD45RA_POS | 63 | 124 | 50.8 |
| Denom = CD4_NEG_CD8_NEG_CD45RA_POS | |||
| LAG3_NEG | 179 | 250 | 71.6 |
| CCR7_NEG | 203 | 250 | 81.2 |
| LAG3_POS | 71 | 250 | 28.4 |
| CCR7_POS | 47 | 250 | 18.8 |
| LAG3_NEG_CCR7_NEG | 132 | 250 | 52.8 |
| LAG3_NEG_CCR7_POS | 47 | 250 | 18.8 |
| CCR7_NEG_LAG3_POS | 71 | 250 | 28.4 |
| LAG3_POS_CCR7_POS | 0 | 250 | 0.0 |
| Denom = CD4_NEG_CD8_POS_CD45RA_POS | |||
| LAG3_NEG | 1367 | 1889 | 72.4 |
| CCR7_NEG | 1370 | 1889 | 72.5 |
| LAG3_POS | 522 | 1889 | 27.6 |
| CCR7_POS | 519 | 1889 | 27.5 |
| LAG3_NEG_CCR7_NEG | 896 | 1889 | 47.4 |
| LAG3_NEG_CCR7_POS | 471 | 1889 | 24.9 |
| CCR7_NEG_LAG3_POS | 474 | 1889 | 25.1 |
| LAG3_POS_CCR7_POS | 48 | 1889 | 2.5 |
| Denom = CD4_POS_CD8_NEG_CD45RA_POS | |||
| LAG3_NEG | 6589 | 6785 | 97.1 |
| CCR7_NEG | 468 | 6785 | 6.9 |
| LAG3_POS | 196 | 6785 | 2.9 |
| CCR7_POS | 6317 | 6785 | 93.1 |
| LAG3_NEG_CCR7_NEG | 384 | 6785 | 5.7 |
| LAG3_NEG_CCR7_POS | 6205 | 6785 | 91.5 |
| CCR7_NEG_LAG3_POS | 84 | 6785 | 1.2 |
| LAG3_POS_CCR7_POS | 112 | 6785 | 1.7 |
| Denom = CD4_POS_CD8_POS_CD45RA_POS | |||
| LAG3_NEG | 113 | 241 | 46.9 |
| CCR7_NEG | 133 | 241 | 55.2 |
| LAG3_POS | 128 | 241 | 53.1 |
| CCR7_POS | 108 | 241 | 44.8 |
| LAG3_NEG_CCR7_NEG | 68 | 241 | 28.2 |
| LAG3_NEG_CCR7_POS | 45 | 241 | 18.7 |
| CCR7_NEG_LAG3_POS | 65 | 241 | 27.0 |
| LAG3_POS_CCR7_POS | 63 | 241 | 26.1 |
Optional: Adding density gates back to GatingSet
Let’s add the gate for LAG3 of CD4+ and CD8+. This is a good visualization to see all sequential gating steps applied to the sample.
# Grab gate as a numeric
current_gate <-
dens_gates |>
dplyr::filter(cd4_pos_cd8_pos == "cd4_neg_cd8_pos") |>
dplyr::pull(LAG3)
# Apply using gs_add_gating-method and
# We want a boundary gate
openCyto::gs_add_gating_method(
gs,
alias = "lag3_cd8",
pop = "+",
parent = "cd4-cd8+",
dims = "LAG3",
gating_method = "boundary",
gating_args = list(min = current_gate, max = Inf)
)
current_gate <-
dens_gates |>
dplyr::filter(cd4_pos_cd8_pos == "cd4_pos_cd8_neg") |>
dplyr::pull(LAG3)
openCyto::gs_add_gating_method(
gs,
alias = "lag3_cd4",
pop = "+",
parent = "cd4+cd8-",
dims = "LAG3",
gating_method = "boundary",
gating_args = list(min = current_gate, max = Inf)
)
ggcyto::autoplot(gs[[1]])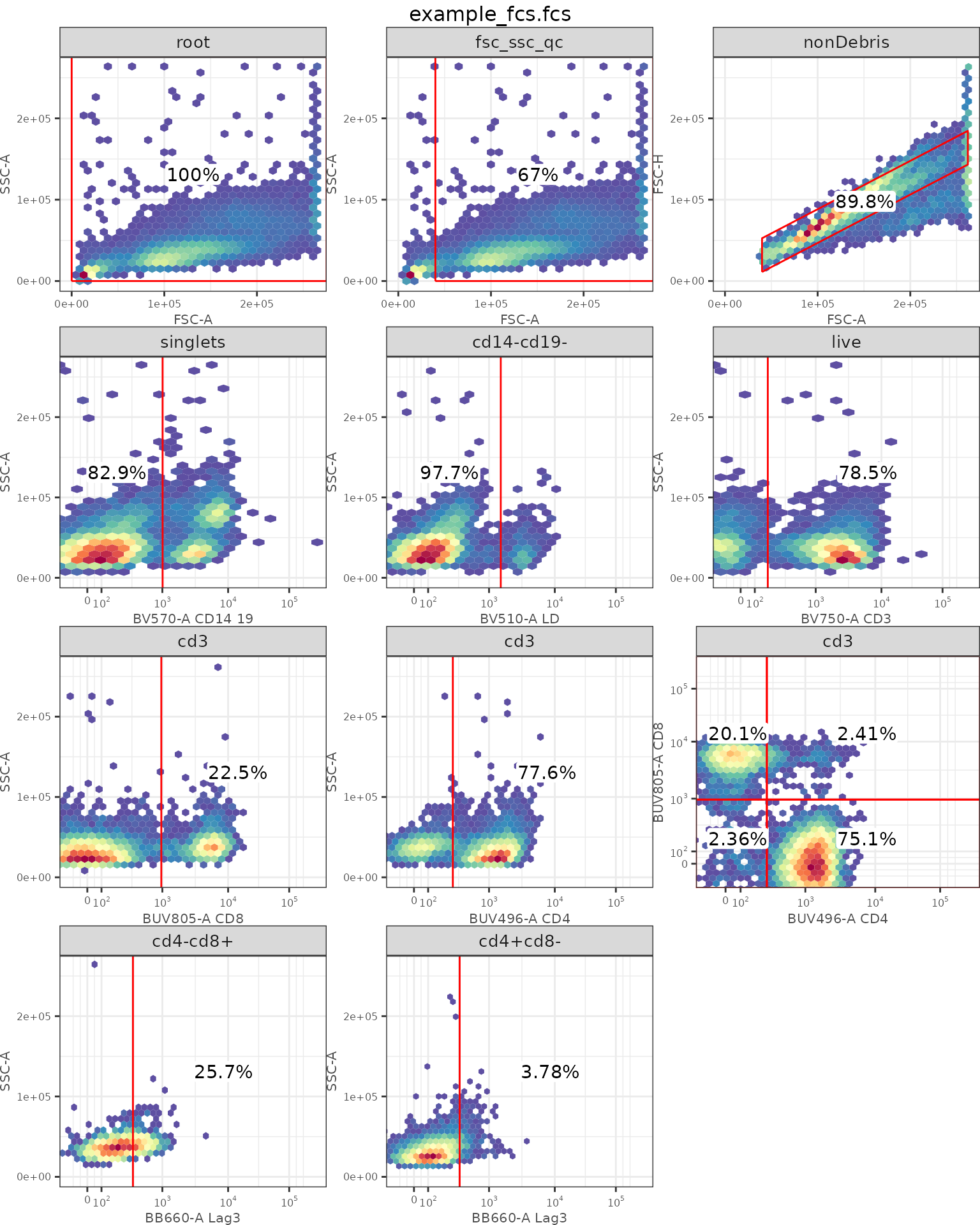
After running {staRgate} to gate flow cytometry data, it is
recommended to perform some quality checks (QC) on the gate placements
to ensure they are reasonable. We suggest to use ridgeplots in addition
to the ggcyto::autoplot function to visualize the density
distributions per marker across samples. When examining a large batch of
samples, downsampling, such as to a random sample of 10k CD3+ cells,
will make the QC process more manageable. In addition, random spot
checks of a few samples would also be helpful QC to detect any edge
cases.
Currently in this tutorial, we do not extend to the QC steps and suggest to lean on other examples for how to put together a ridgeplot for example.
In the near future, we hope to incorporate some examples for the additional QC steps as well, stay tuned!
Session info
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggcyto_1.38.1 ncdfFlow_2.56.0 BH_1.90.0-1
#> [4] ggplot2_4.0.1 flowCore_2.22.1 flowWorkspace_4.22.1
#> [7] openCyto_2.22.0 staRgate_0.99.6 BiocStyle_2.38.0
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.6 xfun_0.56 bslib_0.9.0
#> [4] htmlwidgets_1.6.4 Biobase_2.70.0 lattice_0.22-7
#> [7] vctrs_0.7.0 tools_4.5.2 generics_0.1.4
#> [10] stats4_4.5.2 parallel_4.5.2 tibble_3.3.1
#> [13] pkgconfig_2.0.3 data.table_1.18.0 RColorBrewer_1.1-3
#> [16] S7_0.2.1 desc_1.4.3 S4Vectors_0.48.0
#> [19] gt_1.3.0 graph_1.88.1 lifecycle_1.0.5
#> [22] compiler_4.5.2 farver_2.1.2 stringr_1.6.0
#> [25] textshaping_1.0.4 janitor_2.2.1 snakecase_0.11.1
#> [28] litedown_0.9 htmltools_0.5.9 sass_0.4.10
#> [31] yaml_2.3.12 pillar_1.11.1 pkgdown_2.2.0
#> [34] hexbin_1.28.5 jquerylib_0.1.4 tidyr_1.3.2
#> [37] cachem_1.1.0 RProtoBufLib_2.22.0 commonmark_2.0.0
#> [40] tidyselect_1.2.1 digest_0.6.39 stringi_1.8.7
#> [43] dplyr_1.1.4 purrr_1.2.1 bookdown_0.46
#> [46] labeling_0.4.3 flowClust_3.48.0 fastmap_1.2.0
#> [49] grid_4.5.2 cli_3.6.5 magrittr_2.0.4
#> [52] utf8_1.2.6 RBGL_1.86.0 XML_3.99-0.20
#> [55] withr_3.0.2 scales_1.4.0 timechange_0.3.0
#> [58] lubridate_1.9.4 rmarkdown_2.30 matrixStats_1.5.0
#> [61] gridExtra_2.3 cytolib_2.22.0 ragg_1.5.0
#> [64] evaluate_1.0.5 knitr_1.51 markdown_2.0
#> [67] rlang_1.1.7 Rcpp_1.1.1 glue_1.8.0
#> [70] xml2_1.5.2 Rgraphviz_2.54.0 BiocManager_1.30.27
#> [73] BiocGenerics_0.56.0 jsonlite_2.0.0 R6_2.6.1
#> [76] plyr_1.8.9 systemfonts_1.3.1 fs_1.6.6References
G. Finak, J. Frelinger, W. Jiang, E. W. Newell, J. Ramey, M. M. Davis, S. A. Kalams, S. C. De Rosa and R. Gottardo, “OpenCyto: An Open Source Infrastructure for Scalable, Robust, Reproducible, and Automated, End-to-End Flow Cytometry Data Analysis,” PLoS Computational Biology, vol. 10, p. e1003806, August 2014.
G. Finak and M. Jiang, “flowWorkspace: Infrastructure for representing and interacting with gated and ungated cytometry data sets.,” 2023.
G. Finak, W. Jiang and R. Gottardo, “CytoML for cross-platform cytometry data sharing,” Cytometry Part A, vol. 93, pp. -7, 2018.
G. Monaco, H. Chen, M. Poidinger, J. Chen, J. P. de Magalhães and A. Larbi, “flowAI: automatic and interactive anomaly discerning tools for flow cytometry data,” Bioinformatics, vol. 32, p. 2473–2480, April 2016. P. Van, W. Jiang, R. Gottardo and G. Finak, “ggcyto: Next-generation open-source visualization software for cytometry,” Bioinformatics, 2018.